| Clin Mol Hepatol > Volume 30(1); 2024 > Article |
|
See the commentary-article "Starting the journey: Understanding the roles of complement proteins in liver diseases through mendelian randomization" on page 150.
ABSTRACT
Background/Aims
To evaluate the causal correlation between complement components and non-viral liver diseases and their potential use as druggable targets.
Methods
We conducted Mendelian randomization (MR) to assess the causal role of circulating complements in the risk of non-viral liver diseases. A complement-centric protein interaction network was constructed to explore biological functions and identify potential therapeutic options.
Results
In the MR analysis, genetically predicted levels of complement C1q C chain (C1QC) were positively associated with the risk of autoimmune hepatitis (odds ratio 1.125, 95% confidence interval 1.018ŌĆō1.244), while complement factor H-related protein 5 (CFHR5) was positively associated with the risk of primary sclerosing cholangitis (PSC;1.193, 1.048ŌĆō 1.357). On the other hand, CFHR1 (0.621, 0.497ŌĆō0.776) and CFHR2 (0.824, 0.703ŌĆō0.965) were inversely associated with the risk of alcohol-related cirrhosis. There were also significant inverse associations between C8 gamma chain (C8G) and PSC (0.832, 0.707ŌĆō0.979), as well as the risk of metabolic dysfunction-associated steatotic liver disease (1.167, 1.036ŌĆō1.314). Additionally, C1S (0.111, 0.018ŌĆō0.672), C7 (1.631, 1.190ŌĆō2.236), and CFHR2 (1.279, 1.059ŌĆō1.546) were significantly associated with the risk of hepatocellular carcinoma. Proteins from the complement regulatory networks and various liver diseaserelated proteins share common biological processes. Furthermore, potential therapeutic drugs for various liver diseases were identified through drug repurposing based on the complement regulatory network.
Graphical Abstract
The proportion of non-viral liver diseases among chronic liver diseases (CLD) is increasing, resulting in serious disease and economic burdens [1]. Autoimmune hepatitis (AIH), primary biliary cirrhosis (PBC), and primary sclerosing cholangitis (PSC) represent the three major autoimmune liver diseases [2]. In addition, metabolic dysfunction-associated steatotic liver disease (MASLD) and alcoholic liver disease are two other types of chronic liver disease [3]. MASLD, formerly nonalcoholic fatty liver disease (NAFLD), has been redefined to encompass liver steatosis along with one or more of five specific cardiometabolic risk factors [4]. This renaming and revised criteria reflect a deeper understanding of the diseaseŌĆÖs pathophysiology. Despite the change in name, MASLD continues to exhibit similar prevalence and risk factors as NAFLD, indicating a consistent approach to understanding and managing this liver condition [5-7]. These non-viral liver diseases are anticipated to drive CLD epidemiology going forward and to account for increasing proportions of hepatocellular carcinoma (HCC) and death in the future [8]. There is a lack of effective drugs for the treatment of these non-viral liver diseases, especially when the disease progresses to liver decompensation or HCC, at which point, liver transplantation may be the only effective treatment [9]. Given the lack of effective therapies for non-viral liver diseases and the resulting increase in mortality, it is important to identify biomarkers associated with the occurrence and progression of non-viral liver diseases and to explore their potential as predictive targets for screening therapeutics.
The complement system, a critical component of the innate immune system, is activated by three independent pathways, namely classical, lectin, and alternative, involving more than 50 soluble and membrane-bound proteins [10]. Complement was initially characterized as an essential system for protection against invading pathogens; however, more recent data highlight its role in uncontrolled inflammatory responses that contribute to the development of liver diseases, kidney diseases, tumors, and many other diseases [11]. Most complement proteins are produced in the liver, before being released into the circulation and distributed throughout the body; consequently, changes in the complement system are closely associated with liver diseases [12]. In view of this close association, complement components may be significantly related to the development of liver diseases, and it is of great significance to explore effective complement components as pharmacological intervention targets for the treatment of liver diseases. As the circulating complements are mainly produced by the liver, it is difficult to determine the causal association between the complement components and liver diseases. Whether specific complement components change with the development of liver disease or whether changes in the complement components trigger the occurrence of liver diseases warrants further exploration.
In this study, we developed a new method to investigate the potential causal relationships between complement components and various non-viral liver diseases.
Mendelian randomization (MR) is a powerful genetic epidemiology method that uses human genetics to infer causality. Estimates derived from MR analyses are less susceptible to bias from confounding and reverse causation [13]. Employing MR, we delved into whether changes in these components are a cause or a consequence of risk of developing liver disease. In a pioneering step, we combined our MR findings with a network-based drug repositioning approach, diverging from traditional gene overlap analyses. This method not only identifies potential therapeutic drugs but also strengthens the validity of our MR results. Our study represents the first of its kind to apply MR techniques in evaluating the causal role of complement components in liver diseases and proposes a novel workflow that marries MR insights with network-based drug discovery. These efforts were directed towards filling the gap in effective treatments for non-viral liver diseases, with a particular focus on uncovering new therapeutic targets.
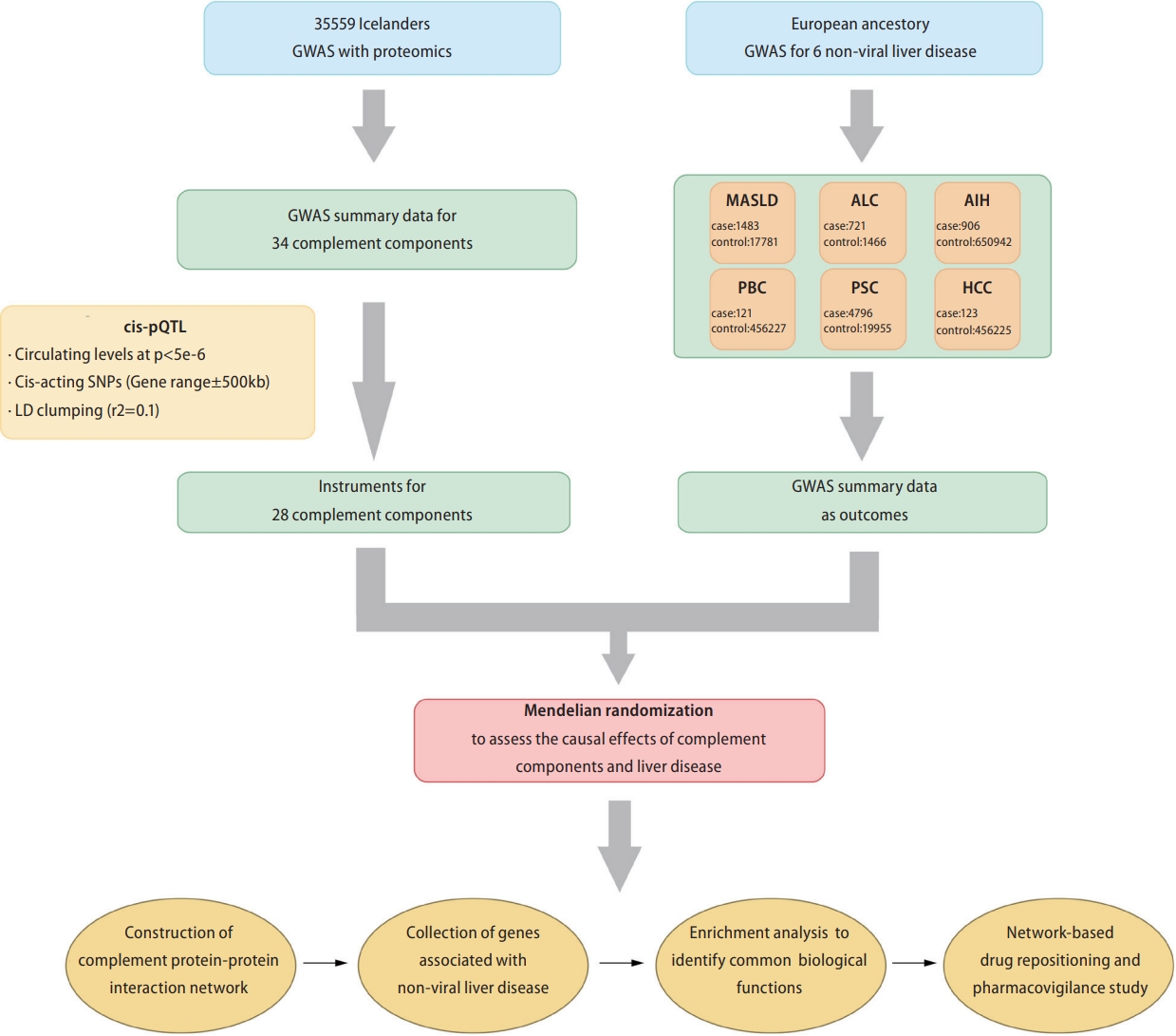
We tested the association between circulating levels of complement proteins with five non-viral liver diseases and HCC using two-sample MR. Subsequently, based on the complement components determined by MR analysis, we constructed a complement protein-protein interaction network for each non-viral liver disease and HCC, explored its involvement in the biological processes of liver diseases, and conducted network-based drug repositioning. Figure 1 shows a schematic representation of the study design. We conducted this study in accordance with the Strengthening the Reporting of Observational Studies in Epidemiology-MR criteria.
Summary data from a proteomic genome-wide association study (GWAS) including 35,559 Icelanders were used to identify cis-acting protein quantitative trait loci (cis-pQTLs) as MR instruments for circulating complements [14]. All 35 complement genes involved in the GWAS were included in our study (Supplementary Table 1). Cis-pQTLs were defined as singlenucleotide polymorphisms (SNPs) located within 500 kb of the gene encoding the measured protein and associated with circulating complement at P<5├Ś10-6. Linkage disequilibrium clumping was performed using the European reference population at an ŌĆ£r2<0.0.1ŌĆØ threshold (window: 10,000 kb). For certain complements (C1q Like 2, C2, Factor B, C1q Binding Protein, C1q Tumor Necrosis Factor [TNF]-Related Protein 1, and C1q Like 4), no significant cis-variants were identified at a threshold of P<5├Ś10-6 (Supplementary Table 2). Therefore, these complements were excluded from further analysis.
Genetic summary data for each non-viral liver disease were obtained from several large population-based cohorts and case-control GWAS. These data sources are listed in Table 1. Detailed information on the summary data for the liver diseases is available in the Supplementary Materials.
We first searched six protein-protein interaction databases (Human Protein Reference Database [https://www.hprd.org/], Interactome INSIDER [http://interactomeinsider.yulab.org/), IntAct [https://www.ebi.ac.uk/intact/], InWeb_IM [http://www.lagelab.org/resources/], PINA [https://omics.bjcancer.org/pina/], BIOGRID [https://thebiogrid.org/], Interactome3D [https://interactome3d.irbbarcelona.org/]) for complement component interacting protein using the Entrez IDs of complement components identified by MR as significantly associated with liver diseases.
Subsequently, we obtained validated complement component interacting proteins by selecting experimentally validated interacting proteins (Supplementary Table 3). Finally, we used the network extension method within the network topology association analysis (NTA), to obtain the top 20 ranking neighboring proteins based on each complement component and its interacting proteins, and constructed a protein interaction network. In the network extension method, random walk analysis was first used to rank all proteins in the selected network based on their network proximity to the input seeds (complement components and their interacting proteins) and then return an expanded subnetwork in which nodes are the input seeds and their top-ranking neighboring proteins and edges represent their relationship. In this study, we used the ŌĆ£WebGestaltRŌĆØ R package (version 0.4.4) to conduct the NTA analysis.
In this study, we obtained liver disease-associated genes from the DisGeNET (https://www.disgenet.org/) [15] and the DISEASES platform (http://diseases.jensenlab.org/) [16]. DisGeNET is a comprehensive discovery platform containing publicly available collections of genes associated with human diseases that integrates data from expert scientific literature, GWAS catalogs, curated repositories, and animal models. The DISEASES platform integrates gene-disease associations from automated text-mining of biomedical literature. We collected 322 alcohol-related cirrhosis (ALC)-associated, 63 MASLD-associated, 191 HCC-associated, 190 AIH-associated, 264 PSCassociated genes, from the DisGeNET and DISEASES platforms (Supplementary Table 4) (accessed in August, 2022).
To identify potential therapeutic drugs for liver diseases, we applied a network proximity approach on the PharmOmics platform (http://mergeomics.research.idre.ucla.edu/runpharmomics.php) [17] to compute the significance of the association between liver disease and drugs. PharmOmics introduces a precise transcriptomic knowledgebase and analytical instrument, enabling drug repositioning through specialized network methodologies for distinct species and tissues. Network-based drug repositioning is based on the connectivity in a given gene network between PharmOmics drug signatures and user input genes such as a disease signature. For each liver disease, the top 15 drugs were selected for pharmacovigilance analysis of their potential hepatotoxicity, and text mining of medical literature and clinical study registries was used to obtain information on the potential therapeutic effects of the predicted drugs on liver disease. Comprehensive information about the construction of a protein-protein interaction network, acquisition of liver diseaseassociated genes, implementation of Gene Ontology (GO) enrichment analysis, identification of drugs with potential hepatotoxicity, and reference bibliographies for evidence of drug repurposing are included in the Supplementary Materials.
We utilized the GTEx Analysis V8 dataset, specifically the ŌĆ£GTEx_Analysis_2017-06-05_v8_RSEMv1.3.0_transcript_ tpm.gct.gzŌĆØ file, to explore the effect of individual complement components on the human liver. Our approach involved analyzing the raw RNA-seq gene ŌĆ£transcripts per millionŌĆØ data from the livers of 226 individuals. To compare liver gene expression, we categorized the expression levels of each complement component into four distinct quartiles. We then focused our analysis on contrasting the gene expression profiles between the highest 25% (upper quartile) and the lowest 25% (lower quartile) of complement component expression levels. This method allowed us to identify significant differences in liver gene expression associated with varying levels of specific complement components.
In our study, we identified differentially expressed genes for enrichment analysis using stringent criteria, specifically a False Discovery Rate (FDR) of < 0.05 and an absolute log Fold Change (|log FC|)Ōēź1.5.
GO biological process and KEGG pathway enrichment analysis was performed using Metascape (https://metascape.org) [18] on the liver diseases-associated genes, proteins from complement components-protein interaction network, and differentially expressed genes related to specific complement components (C1q C chain [C1QC], C1S, complement factor H-related protein [CFHR] 1, CFHR2, CFHR5, C7, and C8 gamma chain [C8G]). The GO biological process and KEGG pathway of co-enrichment of the aforementioned gene sets was demonstrated by heatmaps. This approach allowed us to juxtapose the enriched processes and pathways across the gene sets, emphasizing the specific roles and interactions of complement-related genes in liver diseases.
We performed 165 comparisons and applied a Bonferroni corrected significance level (P<0.05/165=0.0003). Analyses were conducted using the two-sample MR package (version 0.5.6) [19] in R software (version 4.1.2). We use the inverse-variance weighted (IVW) approach as our primary analysis method. Further details regarding the MR methodology are provided in the Supplementary Materials.
AIH, PBC, PSC, MASLD, and HCC GWAS summary statistics are publicly available at ŌĆ£https://www.ebi.ac.uk/gwas/ŌĆØ (last accessed on July 7, 2022) using PMIDs.
Summary statistics for ALC are available at ŌĆ£http://gengastro.med.tu-dresden.de/suppl/alc_cirrhosis/ŌĆØ. Decoded proteomic GWAS summary data can be downloaded at ŌĆ£https://www.decode.com/summarydata/ŌĆØ. We have placed a larger table in the DataS1 file including instrument variables used in the MR analysis, complement components-protein interaction network, liver diseases-associated genes and pharmacovigilance studies using the FDA database.
We obtained GWAS summary data for 35 complements from the deCODE study [14]. A total of 28 complements remained after cis-pQTL definition screening and were included in the MR analysis (Table ). The F-statistics calculated for the instruments of complements analyzed in this study were >10, indicating a low susceptibility to weak instrument bias. The instrumental variables utilized in the MR analysis are listed in Supplementary Table 2.
Genetically predicted C1QC was nominally associated with AIH (IVW, odds ratio [OR]: 1.125, 95% confidence interval [CI] 1.018ŌĆō1.244; P=0.021) based on the cis-pQTL instrument definition (Fig. 2). However, this relationship was not robust in the weighted median (Fig. 3A, B). Based on the proteinŌĆōprotein interaction databases, we obtained a total of eight C1QCinteracting proteins, which were supported by experimental evidence. AIH-associated proteins and those from the C1QC regulatory network were found to be enriched in common biological processes primarily involving inflammation and immune-related processes (Fig. 4A). Figure 4B shows the top 15 potential therapeutic drugs obtained by drug repurposing based on the C1QC regulatory network. Among these drugs, 10 were found to have no hepatotoxicity (Supplementary Table 5), and eight (pioglitazone, medroxyprogesterone, dexamethasone, paricalcitol, losartan, estradiol, calcitriol, and rimonabant) have demonstrated therapeutic effects in various liver diseases in experimental studies. Furthermore, five drugs (pioglitazone, medroxyprogesterone, dexamethasone, calcitriol, and rimonabant) have demonstrated therapeutic effects against liver diseases in clinical studies. Previous studies have also highlighted the therapeutic effects of dexamethasone and calcitriol on AIH (Supplementary Table 6) [20-22].
In accordance with the cis-pQTL definition, a nominally significant association was observed between genetically predicted C8G (IVW, OR: 0.832, 95% CI 0.707ŌĆō0.979; P=0.027) and CFHR5 (IVW, OR: 1.193, 95% CI 1.048ŌĆō1.357; P=0.007) in relation to the risk of PSC (Fig. 2). Although the results from the weighted median were not significant, they were consistent in direction (Fig. 3A, B). Based on proteinŌĆōprotein interaction databases, we identified 13 CFHR5ŌĆōC8G-interacting proteins, which were supported by experimental evidence. PSC-associated proteins and those from the CFHR5ŌĆōC8G regulatory network were found to be enriched in common biological processes, primarily involving the immune response, cell death, stress response, and other processes (Fig. 4C). Figure 4D shows the top 15 potential therapeutic drugs obtained by drug repurposing based on the CFHR5ŌĆōC8G regulatory network. Among these drugs, 12 were found to have no hepatotoxicity (Supplementary Table 5), five (fluoxetine, mycophenolate, dexamethasone, calcitriol, and verteporfin) were found to have therapeutic effects on various liver diseases in experimental studies, and five (dexamethasone, iodine, calcitriol, mycophenolate, and atorvastatin) were found to have therapeutic effects on liver diseases in clinical studies; among these, three drugs (dexamethasone, mycophenolate, and atorvastatin) were found to have therapeutic effects on PSC in a previous study (Supplementary Table 7) [23,24].
We observed no significant association between the 28 genetically predicted complement components and PBC using the IVW method (Fig. 2). Thus, our findings provide limited evidence to support a causal association between the complement system and PBC.
Based on cis-pQTL instrument selection criteria, we observed a nominally significant association between genetically predicted concentrations of C8G and MASLD (IVW, OR: 1.167, 95% CI 1.036ŌĆō1.314; P=0.011, Fig. 2 and Fig. 3A, B). No pleiotropic effects were identified using the MR-PRESSO global test (P=0.732). However, this relationship did not survive the B onfer roni tes t for multip le cor re c tions (P<0.05/165=0.0003). The top three biological processes enriched in proteins from the MASLD and C8G regulatory networks are shown in Figure 4E. Regulation of hormone levels (GO: 0 010 817), c arb ohydrate met ab olic pro cesses (GO:0006109), and lipid metabolic processes (GO:0019216) were the three most common pathways. Among the 10 drugs without hepatotoxicity (Supplementary Table 5), seven (imatinib, thalidomide, verteporfin, thrombin, dexamethasone, erlotinib, and vitamin A) were found to have therapeutic effects on MASLD, one (verteporfin) was found to have effects on HCC in experimental studies, and two (erlotinib and vitamin A) were found to have therapeutic effects on other liver diseases in clinical studies (Fig. 4F, Supplementary Table 8) [25,26].
The genetically predicted CFHR1 concentration was inversely correlated with ALC after controlling for multiple testing (IVW, OR: 0.621, 95% CI 0.497ŌĆō0.776; P=2.74├Ś10-5, Fig. 2); this association was consistent across sensitivity analyses (Fig. 3A, B). We also found nominally significant inverse associations between CFHR2 and ALC (IVW, OR: 0.824, 95% CI 0.703ŌĆō 0.965; P=0.017). MR-PRESSO revealed no evidence of horizontal or outlier variants in these relationships (Fig. 3A, B).
ALC-associated proteins and those from the CFHR1ŌĆō CFHR2ŌĆōC1QC regulatory network were found to be enriched in common biological processes primarily involving inflammation and immune-related processes (Fig. 5A). Figure 5B shows the top 15 potential therapeutic drugs obtained by drug repurposing based on the CFHR1ŌĆōCFHR2ŌĆōC1QC regulatory network. Among these drugs, four were found to have no hepatotoxicity (Supplementary Table 5), four (lenalidomide, losartan, paricalcitol, and bortezomib) were found to have therapeutic effects on other liver diseases in experimental studies, and five (lenalidomide, losartan, decitabine, ascorbic acid, and thalidomide) were found to have therapeutic effects on liver diseases in clinical studies; thalidomide and ascorbic acid were found to have therapeutic effects on ALC in a previous study (Supplementary Table 9) [27,28].
Based on the cis-pQTL definition, CFHR2 (IVW, OR: 1.279, 95% CI 1.059ŌĆō1.546; P=0.011) and C7 (IVW, OR: 1.631, 95% CI 1.190ŌĆō2.236; P=0.002) were positively associated with HCC (Fig. 2 and Fig. 3A, B). HCC-associated proteins and those from the C1sŌĆōCFHR2ŌĆōC7 regulatory network were found to be enriched in common biological processes, primarily immune response, cell activation, stress response, and other processes (Fig. 5C). Six drugs (docetaxel, melphalan, calcitriol, furosemide, thalidomide, and somatostatin) were found to have therapeutic effects on various liver diseases in clinical studies, of which four (docetaxel, melphalan, somatostatin, and thalidomide) were found to have therapeutic effects on HCC (Fig. 5D, Supplementary Table 10) [29,30].
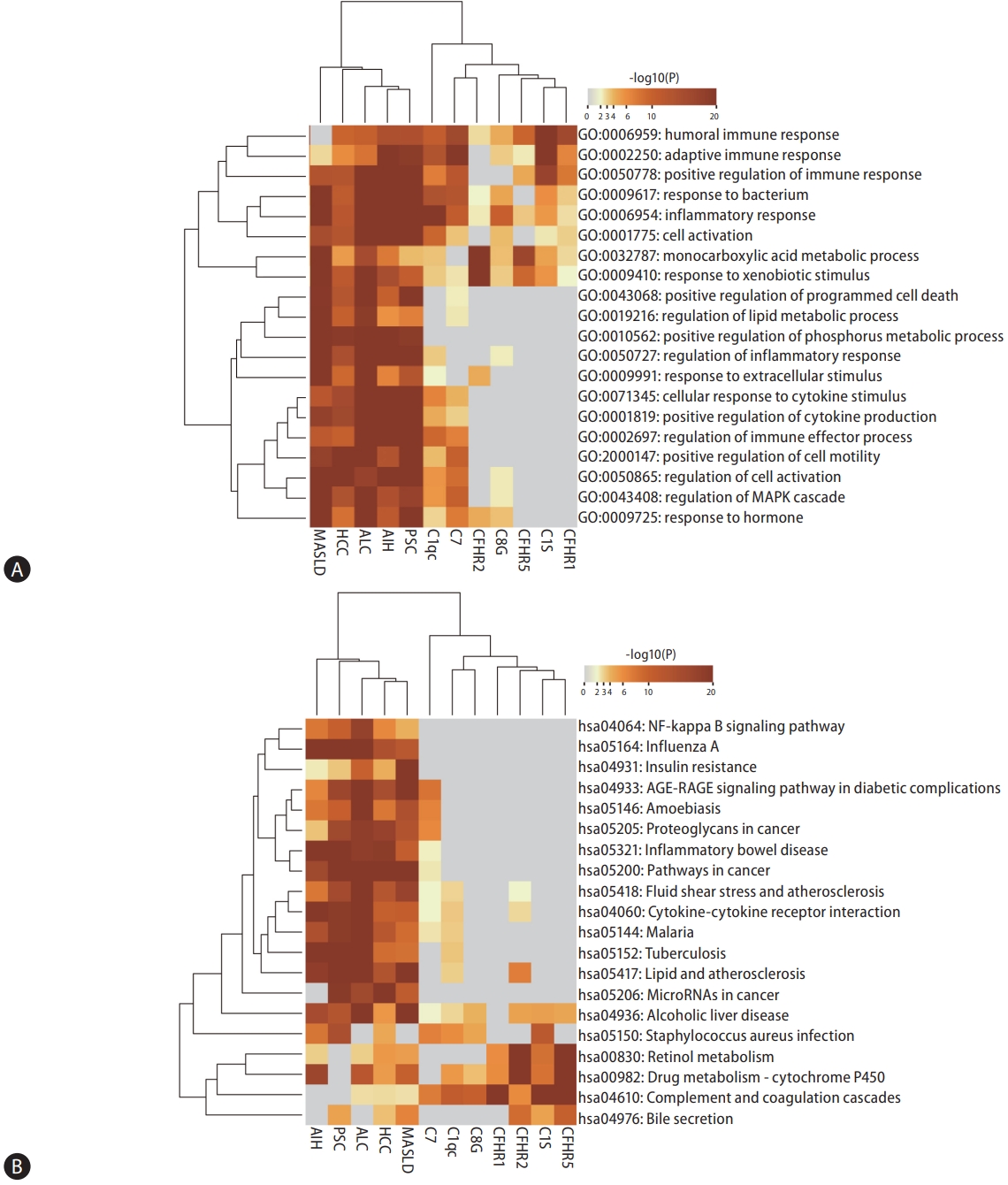
By individually analyzing the expression levels of C1QC, C1S, CFHR1, CFHR2, CFHR5, C7, and C8G, we identified distinct sets of differentially expressed genes for each complement component, totaling 429, 120, 38, 204, 81, 178, and 53 genes, respectively (Supplementary Fig. 1 and 2). In our subsequent enrichment analysis of these genes in the context of nonviral liver diseases, we discovered a significant enrichment in biological processes primarily associated with inflammatory response, metabolic processes, and cell activation (Fig. 6A). Moreover, our analysis revealed that the enriched KEGG pathways predominantly involve complement pathways, drug metabolism, steroid hormone biosynthesis, and retinol metabolism (Fig. 6B). This comprehensive analysis underscores the multifaceted role of complement components in the pathophysiology of nonviral liver diseases, highlighting key areas for potential therapeutic intervention.
In this study, we used cis-MR analyses to evaluate the causal association between genetically predicted circulating levels of complements and non-viral liver diseases. Our results suggest a possible causal relationship between specific complement components and these liver diseases. For example, C1QC may be involved in the development of AIH and ALC. Additionally, C8G and CFHR5 may be associated with PSC, while C8G could potentially be linked to MASLD. Moreover, CFHR1 and CFHR2 might play a role in ALC, whereas C1S, C7, and CFHR2 may be associated with HCC. These findings provide insights into the potential involvement of the complement system in the development of these liver diseases. In addition, we identified several common pathways between proteins from the complement regulatory network and liver disease-related proteins, indicating potential interactions between the complement system and liver diseases. Furthermore, potential therapeutic drugs for various liver diseases have been identified through drug repurposing, utilizing the complement regulatory network.
C1QC was found to be related to the risk of AIH. C1QC, a component of the classical complement pathway, plays a crucial role in immune system regulation. A recent study has found a significant upregulation of C1QC in individuals with AIH when compared to control subjects, further emphasizing its potential importance in the pathogenesis of AIH [31]. Additionally, recent research points to a significant association of C1QC with the progression of colon carcinoma and its involvement in skin cutaneous melanoma. Elevated expression of C1QC in colon neoplasms correlates with a more severe prognosis for affected patients. In a similar vein, C1QC has emerged as an independent predictive factor for the prognosis in cutaneous melanoma [32,33]. Based on the regulatory network of C1QC, we identified a range of potential therapeutic agents for AIH including calcitriol, dexamethasone, medroxyprogesterone, pioglitazone and rimonabant. Notably, the use of calcitriol and dexamethasone is directly supported by experimental or clinical evidence for their effectiveness in AIH. Dexamethasone, a corticosteroid, is widely recognized as the standard treatment for AIH due to its potent anti-inflammatory and immunosuppressive effects [20]. Calcitriol, the active form of vitamin D, has previously been recognized for its immunomodulatory properties [21], which may be pivotal in modulating the autoimmune response in AIH. Patients with chronic-onset AIH exhibit significantly lower vitamin D [22], underscoring the potential benefits of calcitriol supplementation.
We found that CFHR5 expression was positively associated with the risk of PSC, whereas C8G was inversely correlated. CFHR5, a member of the CFHR family, has previously been implicated in other autoimmune and inflammatory conditions, such as glomerulonephritis and systemic lupus erythematosus [34,35]. Among the patients with glomerulonephritis studied, 12.6% (14 individuals) presented with eight distinct exonic variations in the CFHR5 gene. Notably, these patients had lower serum CFHR-5 levels than the control participants. Its role in the complement pathway, which is integral to the innate immune response, suggests a potential mechanism through which it could influence the pathogenesis of PSC. On the other hand, the inverse relationship observed between C8G and PSC risk is equally noteworthy. C8G is one of the three subunits of C8. A study has identified C8G as a potential inhibitor of neuroinflammation [36]. Moreover, the evidence suggests a vital role for C8G in astrocytes for the preservation of the blood-brain barrier during instances of inflammation within the brain [37]. This anti-inflammatory property provides insights into its potential as drug target in PSC. In the drug repurposing analysis, we identified dexamethasone, atorvastatin, calcitriol, iodine, mycophenolate as potential therapeutic agents based on C8G-CFHR5 regulatory network. Among these drugs, dexamethasone and calcitriol show stronger evidence for potential effect in PSC. Specifically, a cohort study of 21 patients treated with 9 mg of budesonide, a corticosteroid like dexamethasone, for 1 year showed an improvement in portal inflammation and a notable reduction in serum alkaline phosphatase and aspartate transaminase levels compared to their baseline [23]. Additionally, in a cholestasis mouse model, the intervention of calcitriol significantly modified gene expression associated with liver bile acid synthesis and transportation, as well as the mRNA expression of proinflammatory cytokines [24].
We found a nominally significant association between the circulating concentrations of C8G and MASLD. Intriguingly, this association was inverted in cases of PSC. MASLD is marked by the accumulation of fat in the liver, whereas PSC is a chronic condition characterized by inflammation and scarring of the bile ducts. This suggests that C8G might play varied roles in these two distinct liver conditions. Moreover, 6 drugs in our drug repositioning analysis showed potential therapeutic effects for MASLD, as supported by existing literature. Vitamin A has shown potential in managing MASLD by influencing lipid metabolism and reducing liver inflammation [25]. On the other hand, Verteporfin, known for its use in photodynamic therapy, might offer benefits in MASLD treatment by targeting pathways involved in liver fibrosis, a complication of the disease [26].
The genetically proxied concentration of CFHR1 and CFHR2 have shown an inverse association with ALC. CFHR1 and CFHR2 belongs to the factor H protein family, which plays a role in regulating innate immune complement reactions, particularly in preventing complement activation by inhibiting C5 convertase and terminal complement complex formation [38]. The activation of C5 contributes to ethanol-induced fatty liver in mice by increasing inflammation [39]. Therefore, it is conceivable that increased circulating concentrations of CFHR1 and CFHR2 may prevent people from developing ALC by inhibiting complement activation. Furthermore, studies have demonstrated that CFHR1 has relevance in malignancies such as lung adenocarcinoma and myelogenous leukemia [40,41]. Specifically, lung adenocarcinoma samples exhibit low levels of CFHR1 and deletion of both alleles of the CFHR1 gene has been identified as a potential predictor for the risk of developing acute myelogenous leukemia. Additionally, our results indicate an association between C1QC and ALC risk. Studies have reported that C1Q-deficient mice displayed a notable resistance to ethanol-induced liver damage, underscored by a decrease in hepatic triglyceride accumulation and a reduction in hepatocellular apoptosis [42]. In our drug repurposing study centered on the CFHR1-CFHR2-C1QC regulator y net work , we identif ied four potential drugs: lenalidomide, losartan, ascorbic acid, and thalidomide. Notably, thalidomide has been reported to prevents alcoholic liver injury in rats [27] and effectively lower the hepatic venous pressure gradient in patients with stable alcoholic cirrhosis, owing to its ability to inhibit the production of TNF-alpha production [28].
We also found that C7 was associated with an increased risk of HCC. C7 plays a crucial role in the lytic pathway of the complement system, leading to the formation of the membrane attack complex. C7 has been recently acknowledged as a novel risk gene for AlzheimerŌĆÖs disease in the Han Chinese population, with the p.K420Q mutation associated with disrupted immune activation and consequent changes in cerebral architecture and functionality [43]. Additionally, C7 is found to be required to maintain the stemness of liver cancer cells. Consistent with our findings, C7 has been previously reported to be upregulated in liver tumor-initiating cells, and depletion of C7 has been found to abrogate the formation of tumor spheres [44]. CFHR2 was found to be associated with HCC. In contrast, a negative correlation was observed between C1S and HCC. C1S is a component of C1, which serves as the primary component of the classical pathway in the complement system. Interestingly, 8 of the 11 drugs without hepatotoxicity were found to have evidence of therapeutic effects on HCC in C1S-CFHR2-C7 network-based drug repurposing. Docetaxel, melphalan, somatostatin, and thalidomide have demonstrated potential therapeutic efficacy against HCC in both basic research and preclinical studies. For example, the results of a phase II trial of thalidomide indicated that it can provide disease stabilization and improve survival rates in patients with HCC [29]. Moreover, a recent study showed that thalidomide inhibits angiogenesis induced by chemotherapy-injured HCC cells [30].
This is the first study using MR techniques to evaluate whether complement components play a causal role in several liver diseases. A strength of our study was that we used variants in close proximity to the encoding gene to minimize horizontal pleiotropy. Pleiotropy observed in cis-SNPs tends to be vertical rather than horizontal, whereas vertical pleiotropy does not violate MR assumptions [45]. To ensure adequate statistical power for our MR analysis, we used summary data from the largest available liver disease GWAS database. We also employed network-based drug repositioning to identify potential therapeutic drugs, which demonstrated stronger and more robust performance than gene overlap-based analysis. Furthermore, we proposed a novel workflow that combines MR findings with network-based drug repositioning to identify potential drugs and confirm the rationality of the MR results.
Our study had several limitations that warrant discussion. After multiple-testing corrections, most of the associations were not preserved in our MR analysis; therefore, the results need to be validated in a GWAS with a larger sample size. Moreover, to reduce the potential population stratification bias, we restricted the GWAS summary datasets to individuals of European ancestry. Hence, the results may not be generalizable to individuals with other ancestries. Despite uncovering substantial experimental and clinical evidence supporting the predicted drugs, there is a pressing need for more extensive research to thoroughly validate these findings. To unlock the full potential of these identified drugs for novel therapeutic applications, a cohesive and efficient research strategy is essential. This should start with detailed preclinical studies to elucidate the drugsŌĆÖ mechanisms and identify any potential adverse effects. Following this, initial clinical trials are crucial to establish safety profiles and optimal dosages. Once these foundational aspects are in place, it becomes imperative to conduct larger-scale clinical trials encompassing a varied patient demographic to ensure a comprehensive evaluation of the drugsŌĆÖ effectiveness. Such a methodical approach is vital for advancing these drugs from promising candidates to viable medical treatments.
In summary, our study provides evidence supporting a causal link between various complement components and liver diseases. These findings suggest that circulating complement components contribute to the pathophysiology of liver diseases and highlight their potential usefulness in drug repositioning.
ACKNOWLEDGMENTS
The scientific calculations in this paper have been done on the HPC Cloud Platform of Shandong University.
This work was supported by the National Natural Science Foundation (Grants No. 82200659), and the Natural Science Foundation of Shandong Province (Grant No. ZR20220H002).
FOOTNOTES
AuthorsŌĆÖ contribution
Conceptualization: CX, XF, GY, YS, XW. Data curation: CX, XF, GY, YS, HD, SS. Formal analysis: XF, YS, JH, JF, JZ, ZL, LL, WW. Funding acquisition: CX, XF, GY. Investigation: YS, HD, ZL, HW, LL, YT. Methodology: CX, XF, GY, YS, JF, JH. Project administration: CX, XF, GY, YS. Resources: CX, XF, GY, YS. Software: XF, YS, HD, SS, XW. Supervision: CX, XF, GY, YS. Validation: CX, XF, GY, YS, HD, SS. Visualization: CX, XF, GY, YS, HD, SS, XW, JF, JZ. Writing ŌĆō original draft: XF, YS, HD. Writing ŌĆō review and editing: CX, XF, GY, YS, HW, WW, YT. All the authors approved this version of the manuscript to be published.
SUPPLEMENTAL MATERIAL
Supplementary material is available at Clinical and Molecular Hepatology website (http://www.e-cmh.org).
Supplementary┬ĀTable┬Ā1.
Complements involved in the GWAS of Iceland
Supplementary┬ĀTable┬Ā2.
Instrument variables of complements used in Mendelian randomization analysis
Supplementary┬ĀTable┬Ā3.
Complement components-protein interaction network
Supplementary┬ĀTable┬Ā5.
Pharmacovigilance studies using the FDA adverse event reporting system
Supplementary┬ĀTable┬Ā6.
Summary of drug repositioning for AIH and supporting evidence
Supplementary┬ĀTable┬Ā7.
Summary of drug repositioning for PSC and supporting evidence
Supplementary┬ĀTable┬Ā8.
Summary of drug repositioning for MASLD and supporting evidence
Supplementary┬ĀTable┬Ā9.
Summary of drug repositioning for ALC and supporting evidence
Supplementary┬ĀTable┬Ā10.
Summary of drug repositioning for HCC and supporting evidence
Supplementary┬ĀFig.┬Ā1.
Enrichment analysis of differential gene expression associated with complement components (C1QC, C1S, and CFHR1) in the context of liver diseases.(A) Distribution of C1QC expression levels (log10(TPM)) in 226 human liver tissues from the GTEx database. The expression levels are categorized into two groups for comparative analysis: the top 25% (high expression, shown in red) and the bottom 25% (low expression, shown in blue). (B, E, G) Volcano plots for differential gene expression in liver tissues associated with varying expression levels of C1QC (B), C1S (E), and CFHR1 (G). Each plot represents genes as points, plotted based on their log fold change (log FC) on the x-axis and the negative log10 of the FDR on the y-axis. Genes meeting the criteria of FDR<0.05 and |log FC|>1 are highlighted: upregulated genes in red and downregulated genes in blue. The top 25 significantly upregulated and downregulated genes are labeled for identification. (C, D, F, H) Heatmaps analyzing GO biological process enrichment in gene sets associated with liver diseases, correlated with differential gene expression in the liver relative to the levels of C1QC (C, D), C1S (F), and CFHR1 (H). Heatmap C details enrichment for AIH in relation to C1QC, D for ALC in relation to C1QC, F for HCC in relation to C1S, and H for ALC in relation to CFHR1. Each heatmap employs a color gradient to represent the significance of enrichment, elucidating the biological processes significantly affected by these liver diseases in the context of complement component levels. GO, gene ontology; AIH, autoimmune hepatitis; ALC, alcohol-related cirrhosis; HCC, hepatocellular carcinoma; C1QC, complement C1q subcomponent subunit C; C1S, complement C1s subcomponent; CFHR1, complement factor H-related protein 1.
Supplementary┬ĀFig.┬Ā2.
Enrichment analysis of differential gene expression associated with complement components (CFHR2, CFHR5, C7, and C8G) in the context of liver diseases. (A, D, F, H) Volcano plots for differential gene expression in liver tissues associated with varying expression levels of CFHR2 (A), CFHR5 (D), C7 (F), and C8G (H). Each plot represents genes as points, plotted based on their log fold change (log FC) on the x-axis and the negative log10 of the false discovery rate (FDR) on the y-axis. Genes meeting the criteria of FDR<0.05 and |log FC|>1 are highlighted: upregulated genes in red and downregulated genes in blue. The top 25 significantly upregulated and downregulated genes are labeled for identification. (B, C, E, G, I, J) Heatmaps analyzing GO biological process enrichment in gene sets associated with liver diseases, correlated with differential gene expression in the liver relative to the levels of CFHR2 (B, C), CFHR5 (E), C7 (G), and C8G (I, J). Heatmap B details enrichment for ALC in relation to CFHR2. C for HCC in relation to CFHR2, E for PSC in relation to CFHR5, G for HCC in relation to C7, I for MASLD in relation to C8G, and J for PSC in relation to C8G. Each heatmap employs a color gradient to represent the significance of enrichment, elucidating the biological processes significantly affected by these liver diseases in the context of complement component levels. GO, gene ontology; AIH, autoimmune hepatitis; PSC, primary sclerosing cholangitis; MASLD, metabolic dysfunction-associated steatotic liver disease; ALC, alcohol- related cirrhosis; HCC, hepatocellular carcinoma; CFHR2, complement factor H-related protein 2, CFHR5, complement factor H-related protein 5; C7, complement component 7; C8G, complement component C8 gamma chain.
Figure┬Ā1.
Schematic of the study design. GWAS, genome-wide association study; MASLD, metabolic dysfunction-associated steatotic liver disease; AIH, autoimmune hepatitis; ALC, alcohol-related cirrhosis; PSC, primary sclerosing cholangitis; PBC, primary biliary cirrhosis; HCC, hepatocellular carcinoma; cis-pQTLs, cis-acting protein quantitative trait loci; SNP, single nucleotide polymorphism.
Figure┬Ā2.
Summary of the IVW results based on the cis-pQTL instruments. Color of tiles are scaled according to MR beta estimates, and white tiles are displayed for associations without available instruments. A single asterisk indicates a nominally significant association (P<0.05), and two asterisks indicate that the association is significant after Bonferroni adjustment for multiple comparisons. IVW, Inverse-variance weighted; cis-pQTLs, cis-acting protein quantitative trait loci; MR, Mendelian randomization; AIH, autoimmune hepatitis; ALC, alcohol-related cirrhosis; PSC, primary sclerosing cholangitis; PBC, primary biliary cirrhosis; MASLD, metabolic dysfunction-associated steatotic liver disease; HCC, hepatocellular carcinoma.
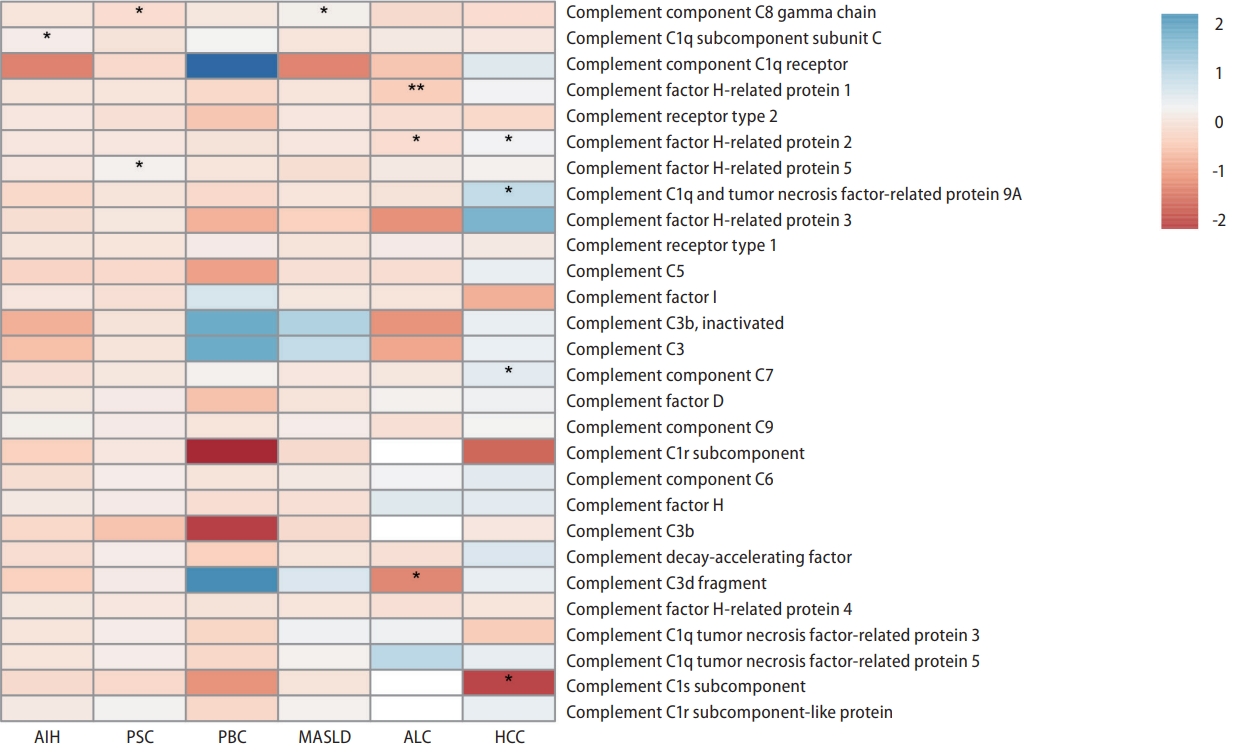
Figure┬Ā3.
Consistent causal relationships and complement pathway visualization. (A) Consistent significant causal relationships estimated using different mendelian randomization (MR) approaches are depicted. Each point on the graph represents the odds ratio (OR) value, while the horizontal lines passing through the points indicate the corresponding 95% confidence interval. The robustness of the observed associations between circulating complement components and various non-viral liver diseases is reinforced by the consistent results across distinct MR techniques. (B) Visualization of the identified associations within the complement pathway, showcasing the dynamic interplay of circulating complement components linked to various non-viral liver diseases. The schematic representation provides an intuitive insight into the alterations of complement components associated with distinct liver diseases. This visualization enhances our understanding of the complex molecular relationships underlying the pathogenesis of non-viral liver diseases. AIH, autoimmune hepatitis; ALC, alcohol-related cirrhosis; PSC, primary sclerosing cholangitis; PBC, primary biliary cirrhosis; MASLD, metabolic dysfunction-associated steatotic liver disease; HCC, hepatocellular carcinoma.
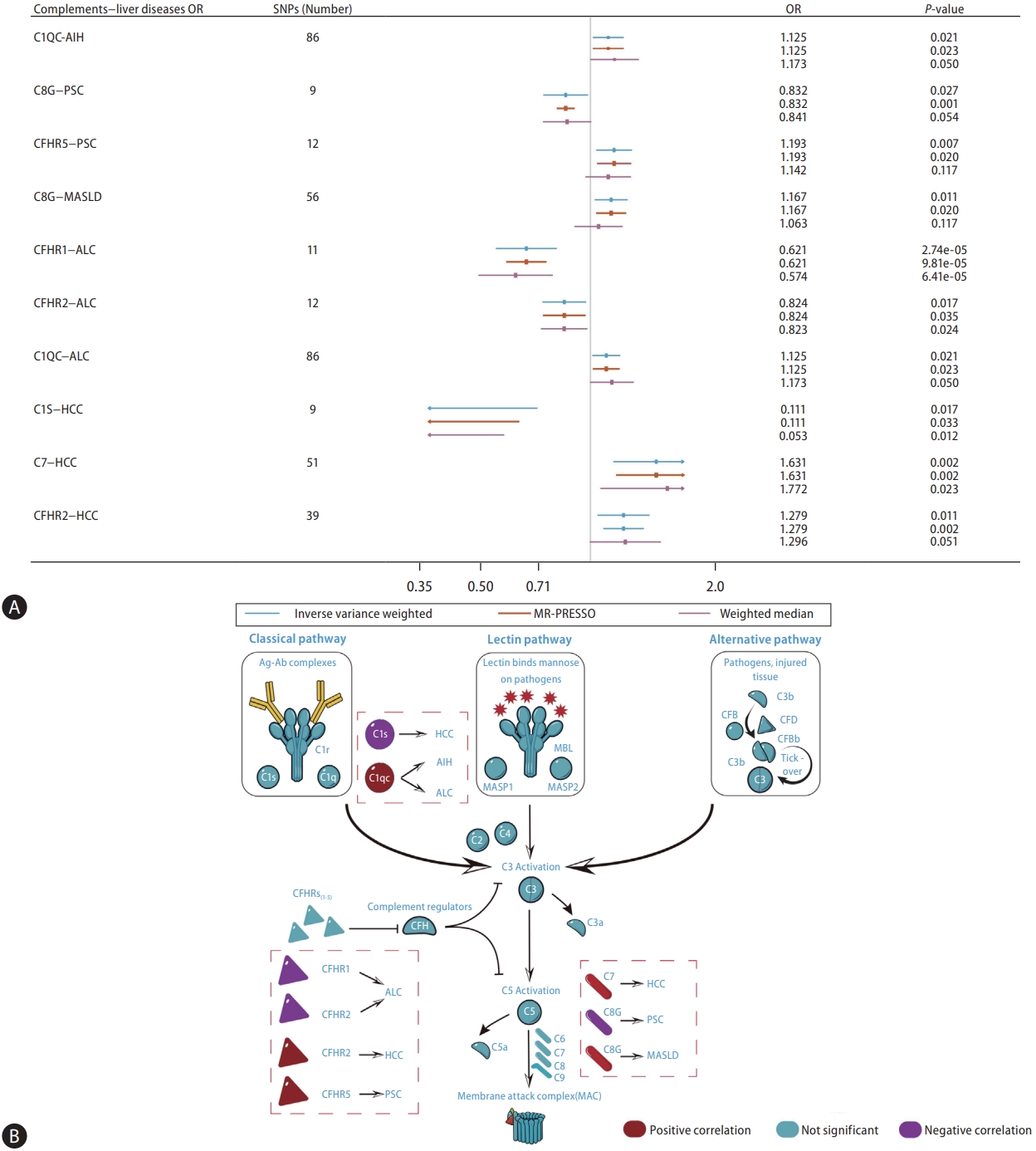
Figure┬Ā4.
Network construction and drug repositioning for AIH, PSC, and MASLD. (A) The top 4 common enriched biological processes identified between the AIH-associated proteins and proteins from the C1QC regulatory network. (B) Sankey diagram illustrating the discovery process of potential therapeutic drugs for AIH based on the C1QC regulatory network through drug repurposing. (C) The top 5 common enriched biological processes identified between the PSC-associated proteins and proteins from the C8G-CFHR5 regulatory network. (D) Sankey diagram illustrating the discovery process of potential therapeutic drugs based on the C8G-CFHR5 regulatory network through drug repurposing. (E) The top 3 common enriched biological processes identified between the MASLD-associated proteins and proteins from the C8G regulatory network. (F) Sankey diagram illustrating the discovery process of potential therapeutic drugs for MASLD based on the C8G regulatory network through drug repurposing. GO, gene ontology; AIH, autoimmune hepatitis; PSC, primary sclerosing cholangitis; MASLD, metabolic dysfunction-associated steatotic liver disease; C1QC, complement C1q subcomponent subunit C; C8G, complement component C8 gamma chain; CFHR5, complement factor H-related protein 5.
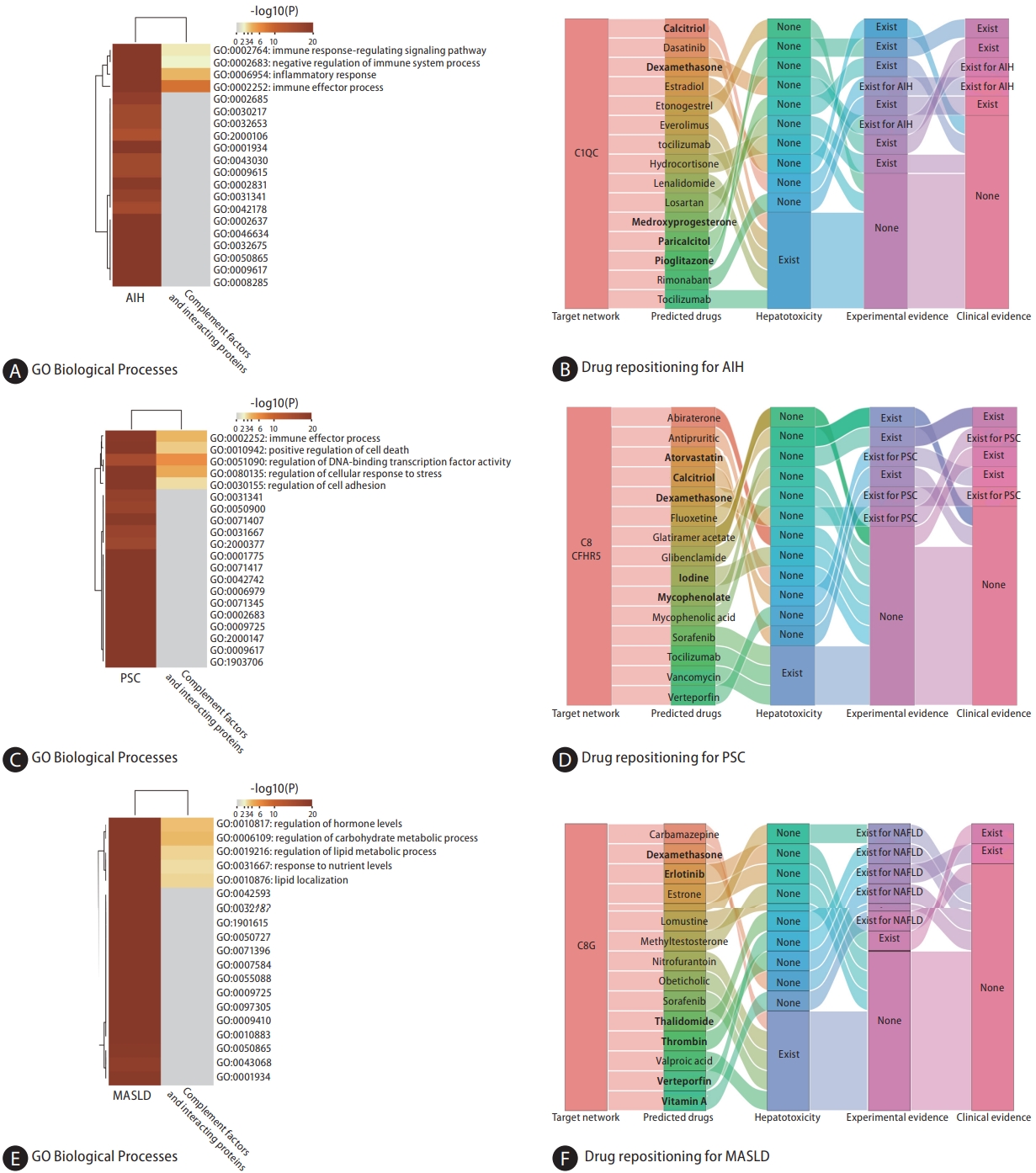
Figure┬Ā5.
Network construction and drug repositioning for ALC and HCC. (A) The top 11 common enriched biological processes identified between the ALC-associated proteins and proteins from the CFHR1-CFHR2-C1QC regulatory network. (B) Sankey diagram illustrating the discovery process of potential therapeutic drugs based on the CFHR1-CFHR2-C1QC regulatory network through drug repurposing. (C) The top 5 common enriched biological processes identified between the HCC-associated proteins and proteins from the C1S-CFHR2-C7 regulatory network. (D) Sankey diagram illustrating the discovery process of potential therapeutic drugs based on the C1S-CFHR2-C7 regulatory network through drug repurposing. GO, gene ontology; ALC, alcohol-related cirrhosis; HCC, hepatocellular carcinoma; C1QC, complement C1q subcomponent subunit C; CFHR1, complement factor H-related protein 1; CFHR2, complement factor H-related protein 2, C1S, complement C1s subcomponent; C7, complement component 7.

Figure┬Ā6.
Enrichment analysis of gene expression linked to complement components in liver diseases. (A) This heatmap offers a detailed analysis of GO biological process enrichment. It focuses on gene sets related to non-viral liver diseases and differential gene expression in the liver, in relation to several complement components (C1QC, C1S, CFHR1, CFHR2, CFHR5, C7, and C8G). The heatmap skillfully visualizes the top 20 enriched term clusters within the GO biological process category, derived from these specific gene sets. (B) This heatmap depicts KEGG pathway enrichment across various input gene lists. It is color-coded to represent P-values, displaying the top 20 clusters of KEGG pathway enrichment. Each cluster is marked by its most significantly enriched pathways, with a nuanced color gradient used to effectively convey the p-value significance. GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; AIH, autoimmune hepatitis; PSC, primary sclerosing cholangitis; MASLD, metabolic dysfunction-associated steatotic liver disease; ALC, alcohol-related cirrhosis; HCC, hepatocellular carcinoma; C1QC, complement C1q subcomponent subunit C; C1S, complement C1s subcomponent; CFHR1, complement factor H-related protein 1; CFHR2, complement factor H-related protein 2, CFHR5, complement factor H-related protein 5; C7, complement component 7; C8G, complement component C8 gamma chain.
Table┬Ā1.
Sources of GWAS summary statistics for the analysis
Table┬Ā2.
Instruments of complements in MR analyses
Abbreviations
AIH
autoimmune hepatitis
ALC
alcohol-related cirrhosis
ALD
alcoholic liver disease
CFHR1
complement factor H-related protein 1
CFHR2
complement factor H-related protein 2
CFHR5
complement factor H-related protein 5
cis-pQTLs
cis-acting protein quantitative trait loci
CLD
chronic liver diseases
C1QC
C1Q subcomponent subunit
C1S
Complement C1s
C7
Complement C7
C8
complement C8
C8G
complement C8 gamma chain
GWAS
genome-wide association study
HCC
hepatocellular carcinoma
GTEx
Genotype-Tissue Expression
GO
Gene Ontology
KEGG
Kyoto Encyclopedia of Genes and Genomes pathway
IVW
inversevariance weighted
MAC
membrane attack complex
MR
mendelian randomization
NAFLD
nonalcoholic fatty liver disease
OR
odds ratio
PSC
primary sclerosing cholangitis
PBC
primary biliary cirrhosis
TICs
tumor-initiating cells
REFERENCES
1. Cheemerla S, Balakrishnan M. Global epidemiology of chronic liver disease. Clin Liver Dis (Hoboken) 2021;17:365-370.




2. Liberal R, Grant CR. Cirrhosis and autoimmune liver disease: Current understanding. World J Hepatol 2016;8:1157-1168.



3. Ikejima K, Kon K, Yamashina S. Nonalcoholic fatty liver disease and alcohol-related liver disease: From clinical aspects to pathophysiological insights. Clin Mol Hepatol 2020;26:728-735.




4. Rinella ME, Lazarus JV, Ratziu V, Francque SM, Sanyal AJ, Kanwal F, et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. J Hepatol 2023;79:1542-1556.

5. Song SJ, Lai JC, Wong GL, Wong VW, Yip TC. Can we use old NAFLD data under the new MASLD definition? J Hepatol 2023 Aug 2. doi: 10.1016/j.jhep.2023.07.021.


6. Ratziu V, Boursier J; AFEF Group for the Study of Liver Fibrosis. Confirmatory biomarker diagnostic studies are not needed when transitioning from NAFLD to MASLD. J Hepatol 2023 Aug 3. doi: 10.1016/j.jhep.2023.07.017.


7. Perazzo H, Pacheco AG, Griep RH; Collaborators. Changing from NAFLD through MAFLD to MASLD: Similar prevalence and risk factors in a large Brazilian cohort. J Hepatol 2023 Sep 9. doi: 10.1016/j.jhep.2023.08.025.


8. Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021;7:6.



9. Yoshiji H, Nagoshi S, Akahane T, Asaoka Y, Ueno Y, Ogawa K, et al. Evidence-based clinical practice guidelines for Liver Cirrhosis 2020. J Gastroenterol 2021;56:593-619.




10. Dunkelberger JR, Song WC. Complement and its role in innate and adaptive immune responses. Cell Res 2010;20:34-50.



11. Ricklin D, Reis ES, Lambris JD. Complement in disease: a defence system turning offensive. Nat Rev Nephrol 2016;12:383-401.




12. Fan X, McCullough RL, Huang E, Bellar A, Kim A, Poulsen KL, et al. Diagnostic and prognostic significance of complement in patients with alcohol-associated hepatitis. Hepatology 2021;73:983-997.


13. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014;23:R89-98.



14. Ferkingstad E, Sulem P, Atlason BA, Sveinbjornsson G, Magnusson MI, Styrmisdottir EL, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet 2021;53:1712-1721.



15. Pi├▒ero J, Ram├Łrez-Anguita JM, Sa├╝ch-Pitarch J, Ronzano F, Centeno E, Sanz F, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res 2020;48:D845-D855.

16. Pletscher-Frankild S, Pallej├Ā A, Tsafou K, Binder JX, Jensen LJ. DISEASES: text mining and data integration of disease-gene associations. Methods 2015;74:83-89.


17. Chen YW, Diamante G, Ding J, Nghiem TX, Yang J, Ha SM, et al. PharmOmics: A species- and tissue-specific drug signature database and gene-network-based drug repositioning tool. iScience 2022;25:104052.



18. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523.




19. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408.


20. Terziroli Beretta-Piccoli B, Mieli-Vergani G, Vergani D. Autoimmune hepatitis: Standard treatment and systematic review of alternative treatments. World J Gastroenterol 2017;23:6030-6048.



21. Prietl B, Treiber G, Pieber TR, Amrein K. Vitamin D and immune function. Nutrients 2013;5:2502-2521.



22. Abe K, Fujita M, Hayashi M, Takahashi A, Ohira H. Association of serum 25-hydroxyvitamin D levels with severe necroinflammatory activity and inflammatory cytokine production in type I autoimmune hepatitis. PLoS One 2020;15:e0239481.



23. Angulo P, Jorgensen RA, Keach JC, Dickson ER, Smith C, Lindor KD. Oral budesonide in the treatment of patients with primary biliary cirrhosis with a suboptimal response to ursodeoxycholic acid. Hepatology 2000;31:318-323.


24. Ogura M, Nishida S, Ishizawa M, Sakurai K, Shimizu M, Matsuo S, et al. Vitamin D3 modulates the expression of bile acid regulatory genes and represses inflammation in bile duct-ligated mice. J Pharmacol Exp Ther 2009;328:564-570.


25. Saeed A, Bartuzi P, Heegsma J, Dekker D, Kloosterhuis N, de Bruin A, et al. Impaired hepatic vitamin A metabolism in NAFLD mice leading to vitamin A accumulation in hepatocytes. Cell Mol Gastroenterol Hepatol 2021;11:309-325.e3.



26. Song K, Kwon H, Han C, Chen W, Zhang J, Ma W, et al. Yesassociated protein in kupffer cells enhances the production of proinflammatory cytokines and promotes the development of nonalcoholic steatohepatitis. Hepatology 2020;72:72-87.




27. Enomoto N, Takei Y, Hirose M, Ikejima K, Miwa H, Kitamura T, et al. Thalidomide prevents alcoholic liver injury in rats through suppression of Kupffer cell sensitization and TNF-alpha production. Gastroenterology 2002;123:291-300.


28. Austin AS, Mahida YR, Clarke D, Ryder SD, Freeman JG. A pilot study to investigate the use of oxpentifylline (pentoxifylline) and thalidomide in portal hypertension secondary to alcoholic cirrhosis. Aliment Pharmacol Ther 2004;19:79-88.



29. Patt YZ, Hassan MM, Lozano RD, Nooka AK, Schnirer II, Zeldis JB, et al. Thalidomide in the treatment of patients with hepatocellular carcinoma: a phase II trial. Cancer 2005;103:749-755.


30. Dong G, Zheng QD, Ma M, Wu SF, Zhang R, Yao RR, et al. Angiogenesis enhanced by treatment damage to hepatocellular carcinoma through the release of GDF15. Cancer Med 2018;7:820-830.


31. Shin E, Schwarz KB, Jones-Brando LV, Florea LD, Sabunciyan S, Wood LD, et al. Expression of HLA and autoimmune pathway genes in liver biopsies of young subjects with autoimmune hepatitis type 1. J Pediatr Gastroenterol Nutr 2022;75:269-275.



32. Deng H, Chen Y, Liu Y, Liu L, Xu R. Complement C1QC as a potential prognostic marker and therapeutic target in colon carcinoma based on single-cell RNA sequencing and immunohistochemical analysis. Bosn J Basic Med Sci 2022;22:912-922.




33. Yang H, Che D, Gu Y, Cao D. Prognostic and immune-related value of complement C1Q (C1QA, C1QB, and C1QC) in skin cutaneous melanoma. Front Genet 2022;13:940306.



34. Garam N, Cserhalmi M, Proh├Īszka Z, Szil├Īgyi ├ü, Veszeli N, Szab├│ E, et al. FHR-5 serum levels and CFHR5 genetic variations in patients with immune complex-mediated membranoproliferative glomerulonephritis and C3-glomerulopathy. Front Immunol 2021;12:720183.


35. Hu X, Liu H, Du J, Chen Y, Yang M, Xie Y, et al. The clinical significance of plasma CFHR 1-5 in lupus nephropathy. Immunobiology 2019;224:339-346.


36. Kim JH, Afridi R, Han J, Jung HG, Kim SC, Hwang EM, et al. Gamma subunit of complement component 8 is a neuroinflammation inhibitor. Brain 2021;144:528-552.



37. Kim JH, Han J, Suk K. Protective effects of complement component 8 gamma against blood-brain barrier breakdown. Front Physiol 2021;12:671250.



38. Heinen S, Hartmann A, Lauer N, Wiehl U, Dahse HM, Schirmer S, et al. Factor H-related protein 1 (CFHR-1) inhibits complement C5 convertase activity and terminal complex formation. Blood 2009;114:2439-2447.



39. Pritchard MT, McMullen MR, Stavitsky AB, Cohen JI, Lin F, Edward Medof M, et al. Differential contributions of C3, C5, and decay-accelerating factor to ethanol-induced fatty liver in mice. Gastroenterology 2007;132:1117-1126.



40. Fratelli M, Bolis M, Kurosaki M, Dori M, Guarnaccia V, Spinelli O, et al. Association of CFHR1 homozygous deletion with acute myelogenous leukemia in the European population. Leuk Lymphoma 2016;57:1234-1237.


41. Wu G, Yan Y, Wang X, Ren X, Chen X, Zeng S, et al. CFHR1 is a potentially downregulated gene in lung adenocarcinoma. Mol Med Rep 2019;20:3642-3648.



42. Cohen JI, Roychowdhury S, McMullen MR, Stavitsky AB, Nagy LE. Complement and alcoholic liver disease: role of C1q in the pathogenesis of ethanol-induced liver injury in mice. Gastroenterology 2010;139:664-674.e1.



43. Zhang DF, Fan Y, Xu M, Wang G, Wang D, Li J, et al. Complement C7 is a novel risk gene for AlzheimerŌĆÖs disease in Han Chinese. Natl Sci Rev 2019;6:257-274.




- TOOLS
-
METRICS

-
- 1 Crossref
- 0 Scopus
- 3,010 View
- 156 Download
- ORCID iDs
-
Guandou Yuan

https://orcid.org/0000-0002-4758-1928Xiude Fan

https://orcid.org/0000-0001-7413-080X - Related articles




 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Supplement1
Supplement1 Print
Print