| Clin Mol Hepatol > Volume 27(1); 2021 > Article |
|
See the commentary-article "Probiotics for treatment of nonalcoholic fatty liver disease: It is worth a try" on page 83.
ABSTRACT
Background/Aims
Nonalcoholic fatty liver disease (NAFLD) is closely related to gut-microbiome. There is a paucity of research on which strains of gut microbiota affect the progression of NAFLD. This study explored the NAFLD-associated microbiome in humans and the role of Lactobacillus in the progression of NAFLD in mice.
Methods
The gut microbiome was analyzed via next-generation sequencing in healthy people (n=37) and NAFLD patients with elevated liver enzymes (n=57). Six-week-old male C57BL/6J mice were separated into six groups (n=10 per group; normal, Western, and four Western diet + strains [109 colony-forming units/g for 8 weeks; L. acidophilus, L. fermentum, L. paracasei, and L. plantarum]). Liver/body weight ratio, liver pathology, serum analysis, and metagenomics in the mice were examined.
Results
Compared to healthy subjects (1.6±4.3), NAFLD patients showed an elevated Firmicutes/Bacteroidetes ratio (25.0±29.0) and a reduced composition of Akkermansia and L. murinus (P<0.05). In the animal experiment, L. acidophilus group was associated with a significant reduction in liver/body weight ratio (5.5±0.4) compared to the Western group (6.2±0.6) (P<0.05). L. acidophilus (41.0±8.6), L. fermentum (44.3±12.6), and L. plantarum (39.0±7.6) groups showed decreased cholesterol levels compared to the Western group (85.7±8.6) (P<0.05). In comparison of steatosis, L. acidophilus (1.9±0.6), L. plantarum (2.4±0.7), and L. paracasei (2.0±0.9) groups showed significant improvement of steatosis compared to the Western group (2.6±0.5) (P<0.05).
Graphical Abstract
Nonalcoholic fatty liver disease (NAFLD) is characterized by liver lipid accumulation or inflammation. Also, it is one of the most common chronic liver diseases and can lead to the development of liver diseases, such as cirrhosis and hepatocellular carcinoma [1]. However, there is not yet an effective treatment method for NAFLD. Recently, fat-related diseases have been on the rise in association with increased consumption of westernized diet. Therefore, future studies exploring new effective treatments for NAFLD are needed.
Gut microbiome plays an important role in human physiology with significant impacts on inflammation and disease [2]. A direct connection between metabolic syndromes and intestinal dysbiosis has been previously established [3]. Intestinal dysbiosis has been shown to be a contributing factor to obesity, insulin resistance, and fatty liver progression [4]. A balanced microbiota is integral to the maintenance and change in intestinal barrier function [5]. In addition, the composition of bacterial species in the gut is unique to the host and has an important role in bacterial translocation, immune system, and vitamin production [6]. Disturbances of gut microbiota have been associated with a number of liver diseases, including NAFLD [7]. Portal vein directly links the liver to the gut, and mediates the transfer of nutrients and microbial components along the gut-liver axis [8]. Therefore, recent studies present a major target for designing novel therapeutics for intestinal inflammatory disorders [9].
A previous study reported that cholesterol is a significant risk factor for NAFLD [10]. Another study demonstrated that cholesterol makes the liver susceptible to fatty hepatitis induced by tumor necrosis factor (TNF)-α and inflammatory pathway in rats [11]. In addition, liver cholesterol synthesis was increased in NAFLD patients, and the regulation of cholesterol metabolism was lowered [12]. These results suggest that elevated cholesterol level is significantly associated with NAFLD.
Lactobacillus has been known to be beneficial in the regulation of immune system and the promotion of anti-inflammatory effects [13-15]. A previous study has demonstrated that probiotics-treatment resulted in reduction of liver steatosis and liver enzyme in NAFLD animal model [16]. Therefore, the modulation of the gut microbiome with probiotics can be considered a strategy for the regulation of metabolic syndrome and obesity-related comorbidities, which include dyslipidemia and insulin resistance. Recent studies are continuously adding to the evidence that supports this theory [17]. In our study, we collected and compared stools from normal people (with body mass index [BMI] 25 kg/m2 or less) and NAFLD patients with elevated liver enzymes. In addition, we demonstrated the role of Lactobacillus acidophilus, L. fermentum, L. paracasei, and L. plantarum strains in the progression of NAFLD in mice.
This prospective observational study was carried out between April 2017 and March 2020 (Table 1). A total of 94 patients comprising of healthy controls (n=36) and NAFLD patients with elevated liver enzyme (n=54) were enrolled and analyzed. The patients underwent standard treatment for their disease, regardless of their enrollment in the study. A healthy control group was collected from health promotion center. Patients who had NAFLD with an elevated level of liver enzyme (aspartate aminotransferase [AST] >50 IU/L, alanine aminotransferase [ALT] >50 IU/L, and BMI score ≥25 kg/m2) were enrolled as NAFLD with elevated liver enzyme group. The exclusion criteria were as follows: patients with a history of viral hepatitis, heavy alcohol drinking within 3 months, alcoholic hepatitis, autoimmune hepatitis, pancreatitis, hemochromatosis, Wilson’s disease, cancer, and drug-induced liver injury. This study was controlled in accordance with ethical guidelines from the 1975 Helsinki Declaration as reflected by prior approval by the institutional review notice for human research in a hospital participating in the trial (2016-134). Basic information has been registered on ClinicalTrials.gov in the public trial registry (NCT04339725). Informed agreement for enrollment was received from each participant.
Baseline evaluation was performed on complete blood count, liver function test, and viral markers. Patients with NAFLD underwent abdominal ultrasound or computed tomography. AST, ALT, creatine, total cholesterol, gamma glutamyl transferase (γGT), triglyceride (TG), fasting blood sugar (FBS), and high density lipoprotein cholesterol were included as serum biochemical parameters. Tests for hepatitis viruses and human immunodeficiency were conducted in all subjects. Enrolled patients and control groups underwent stool sampling and clinical analysis. Clinical data were simultaneously matched with metagenomics data. Fecal samples were obtained in a plastic collection kit at various times during the day. All samples were stored at −80°C. In the case of healthy controls, the patients collected the stool samples at home and kept them at −20°C in a refrigerator. The patients then sent the stool box to the hospital, where the samples were kept at −80°C in a refrigerator.
Metagenomic DNA was extracted using a QIAamp stool kit (cat. no. 51504; QIAGEN, Hilden, Germany). Barcoded universal primers were utilized in the amplification of the V3–V4 region of the bacterial 16S rRNA gene. Polymerase chain reaction (PCR) has conducted in the following steps: denaturation at 95°C for 5 minutes, 20 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, followed by extension at 72°C for 10 minutes. Agencourt AMPure XP system (Beckman Coulter, Pasadena, CA, USA) was employed for the purification of amplicons. PicoGreen and quantitative PCR was utilized for the quantification of the purified amplicons. Following the pooling of the barcoded amplicons, MiSeq sequencer on the Illumina platform (ChunLab Inc., Seoul, Korea) was used for sequencing according to the manufacturer’s specifications.
The 16S-based Microbial Taxonomic Profiling platform of EzBioCloud Apps (ChunLab Inc.) was used for microbiome profiling. Following the taxonomic profiling of each sample, a comparative analysis of the samples was performed by a comparative of EzBioCloud Apps. ChunLab’s 16S rRNA database (DB ver. PKSSU4.0) [18] was used for the taxonomic assignment of reads. Operational taxonomic unit picking was achieved with UCLUST [19] and CDHIT using a 97% similarity cutoff [20]. Beta-diversity, which includes principal coordinates analysis and unweighted pair group method with arithmetic mean clustering, was displayed in the comparative multi touch point (MTP) analyzer. All 16S rRNA were subsequently deposited in the ChunLab’s EzBioCloud Microbiome Database and National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject number PRJNA532302.
Using 16S rRNA sequencing, primers were prepared according to the constant region, and base sequence of the variable region was confirmed through PCR amplification.
L. acidophilus was isolated from the feces of newborn, and L. fermentum was isolated from the saliva of newborn. L. paracasei was sourced from water kimchi and L. plantarum was isolated from spinach. The fecal samples were obtained and isolated via a voluntary donation from the members of the coauthors’ affiliated institutions. And the used Lactobacillus spp. were inoculated into a flask which included de Man, Rogosa and Sharpe medium (MRS) (BD/Difco). The strains were subsequently cultured in anaerobic conditions at 37°C for 24 hours. Stocks of each strain were produced by mixing of the culture broth with an equivalent 20% skim milk solution; they were stored at −80°C until use. The seed culture grew in a flask which contained MRS broth for Lactobacillus spp. at 37°C for 24 hours. Each broth was inoculated in an optimized medium inside a fermenter (MARADO-05D-PS; Bio Control & Science, Daejeon, Korea). The fermentation was performed with a constant pH of 5.5 to 6.0 being maintained by automatically addition of NaOH solution (25% w/v) under 120 rpm agitation at 37°C for 18 to 20 hours. Following fermentation, the cells were harvested via centrifugation at 6,000 rpm for 10 minutes (Supra R12; Hanil, Gimpo, Korea). The lyophilization of 40X enriched cells was performed in accordance with the manual (Lab-Mast 10; Cooling & Heating System, Siheung, Korea). Following lyophilization, colony-forming units (CFU) per gram of each strain powder were measured out via dilution. Strains underwent suspension in 0.1 M phosphate-buffed saline, and were regulated to a density of 109 CFU/mL before use. The reference characteristics of probiotics strains are provided in Table 2.
Six-week old specific-pathogen-free C57BL/6J male mice were sourced from Doo-Yeol Biotech (Seoul, Korea). All mice were housed in individual steel micro isolator cages that were maintained at 22±2°C in a 12/12-hour light/dark cycle. Throughout the experiment, the mice had free access to water and food, and were monitored daily. The experiment design included an adaptation period for all groups, during which the mice were fed a normal diet for a week, and the groups receiving a westernized diet were given a 3-day intake adaptation period. The mice were treated humanely, and all aspects of the animal study was conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. This experiment was designed by selecting a candidate for the treatment of liver disease among the probiotics approved by South Korea’s Ministry of Food and Drug Safety. All of the procedures were licensed by the Institutional Animal Care and Use Committee of the College of Medicine, Hallym University (2018–04).
The male C57BL/6J mice had free access to water and food, and were monitored daily (n=10 mice per each group). The mice were distributed into six groups: normal (n=10), Western diet (n=10), and Western diet with strain groups (n=10 per group). The Western diet (TD88137, Seoul, Korea) was sourced from Doo-Yeol Biotech, and it consisted of 42.7% carbohydrate, 42% fat, and 15% protein. Probiotics were suspended in distilled water maintaining a concentration of 109 CFU/g for 8 weeks. The strains used were L. acidophilus, L. fermentum, L. paracasei, and L. plantarum. The mice were eventually sacrificed via overdose of inhalation anesthesia (isoflurane, Aerane; Baxter, Deerfield, IL, USA) at the conclusion of treatment period. Following the weighing of the mouse blood, liver, feces, and small intestine were collected. Serum was collected via centrifugation (5 minUTES for 19,000 ×g) from whole blood (800 μL). The liver and feces were excised and subsequently stored at –80°C.
From animal serum, the total cholesterol, AST, and ALT were quantified using a biochemical blood analyzer (KoneLab 20; Thermo Fisher Scientific, Waltham, Finland). The AST/ALT ratio is calculated as a method to elucidate the etiology of liver damage [21].
Triglyceride Quantification Kit (Sigma, St-Louis, MO, USA) was used to determine liver TG by colorimetric method. Liver tissue samples were homogenized in 5% Nonidet P 40 Substitute (Roche, Mannheim, Germany). The samples were heated to 100℃ in water bath for 5 minutes or until the Nonidet P 40 became cloudy, and then cooled to room temperature. Heating was reported once more to solubilize all TG. The samples were centrifuged for 2 minutes at maximum speed to remove insoluble material, and then diluted 100-fold with deionized water before assay.
A 10% formalin was used in the fixation of specimens, which were embedded in paraffin. The tissue sections underwent staining with hematoxylin and eosin. The liver was categorized in accordance with the clinical research network scoring system for NAFLD, which ranges from grade 0 to 3 (0: <5%, 1: 5–33%, 2: 34–66%, and 3: >66% of steatosis). Inflammation was categorized according to grades 0 to 3 (0: none, 1: 1–2 foci per ×20 field, 2: 2–4 foci per ×20 field, and 3: >4 foci per ×20 field). A hepato-pathologist (S.H.H.) analyzed all of the biopsy specimens.
Liver tissue samples were homogenized in TRIzol reagent (Invitrogen, Gaithersburg, MD, USA). High Pure RNA Isolation Kit (Roche) was used to isolate RNA from liver tissue. Total RNA isolation from tissue was used a cDNA reverse transcription kit (applied Biosystems, Foster City, CA, USA), and aliquots of total RNA (2 μg) were transformed into cDNA. The cDNA subsequently underwent amplification for quantitative PCR using the Luna® Universal Probe qPCR Master Mix (New England Biolabs, Beverly, MA, USA) and target-specific probe-primer (applied Biosystems).
Continuous variables were expressed as means and standard deviations. One-way analysis of variance (ANOVA) and independent sample T-test were performed for body weight, liver function test, and histological analyses. For additional statistical analysis, data underwent normalization based on MSTUS18, which is implemented in NOREVA (http://idrblab.cn/noreva/). Multiple Experiment Viewer was employed for hierarchical clustering analysis and ANOVA with a post hoc test. P-value <0.05 was considered statistically significant. All statistical analyses were performed using the SPSS software (ver. 19; SPSS Inc., Chicago, IL, USA).
This study was performed between April 2017 and March 2020. A total of 94 patients comprising normal controls (n=37) and NAFLD with elevated liver enzyme (n=57) groups were classified according to their conditions and then analyzed (Fig. 1). The sample providers were all over the age of 19 years, the groups were screened by AST, ALT, and BMI scores. The mean cholesterol level was not significantly different between groups. However, the mean levels of AST, ALT, γGT, TG, and FBS were significantly lower in the normal group (P<0.05). Also, BMI was significantly different between groups (P<0.001) (Table 1).
Bacteroides and Firmicutes are major phylum of domain bacteria that dominate the human gut microbiota (Fig. 2A) [22]. Compared to healthy subjects (1.6±4.3), NAFLD patients showed elevated Firmicutes/Bacteroidetes (F/B) ratio (25.0±29.0), and had reduced composition of Akkermansia and L. murinus (P<0.05). For the comparison of taxonomic composition at the phylum level, Firmicutes increased in the NAFLD with elevated liver enzyme group (51.8%) compared to normal controls (35.1%) (Fig. 2B). We compared the relative abundance of the Lactobacillales family in the normal control and NAFLD groups, and found a statistically significant difference in the level of Aerococcaceae (P<0.001). There was no statistically significant difference in the level of Enterococcaceae, Streptococcaceae, and Lactobacillaceae between the normal control and NAFLD with elevated liver enzyme groups (Fig. 2C). In the genus level, there was a significant difference in the levels of Streptococcus and Granulicatella between the normal control and NAFLD with elevated liver enzyme groups (P<0.05) (Fig. 2D). There were differences in the ratios of L. fermentum, L. paracaseim, and L. plantarum between normal control and NAFLD with elevated liver enzyme groups, however, there was no statistical difference. Also, L. rogosae and L. sakei levels were increased in the NAFLD group (P<0.05) (Fig. 2E).
The mice liver specimen confirmed that NAFLD was induced in the Western diet group (Fig. 3A, B). During 8 weeks of Western diet intake, L. acidophilus group had the lowest weight gain (Fig. 3C). In the analysis of the liver-to-body weight ratio (%), the L. acidophilus (5.5±0.4) group was related to a significant improvement compared to the Western diet group. The L. plantarum (6.0±0.5), L. paracasei (5.7±0.4), and L. fermentum (5.9±0.6) groups did not show a significant difference in the liver-to-body weight ratio (P>0.05) (Fig. 3D).
Several biochemical tests were performed to evaluate the liver function status. In the liver function test, the Western diet group showed remarkably increased liver enzyme (AST) levels compared to the normal control group (P<0.05). The mean levels of AST and ALT in the strain groups were lower than that of the Western diet group, but there is no significant difference (P>0.05). All of the strains except L. paracasei caused a significant decrease in the serum total cholesterol level compared to the Western diet group (P<0.05) (Fig. 4A). Steatosis grade in L. paracasei, L. plantarum, and L. acidophilus groups were lower compared to the Western diet control group (P<0.05) (Fig. 4B). Regarding steatosis scores, all of the Western diet groups had a score of almost 3, whereas the strain groups generally had scores in the early 2 range. Moreover, large fat vacuoles displaced the nuclei to the edges of the cells, representing minimal to mild mixed macro/microvascular steatosis. Only L. fermentum showed no significant meaning in the comparison of steatosis scores (Fig. 4C).
Triglyceride is a type of lipid that circulate in blood. In the analysis of the liver TG level (ng/µL), all of the groups with the strain L. acidophilus (800.8±135.2), L. fermentum (930.9±173.2), L. paracasei (770.7±289.6), and L. plantarum (1,305.6±213.3) showed an improvement compared to the Western diet group (1,446.2±252.2). Except for L. plantarum, all of the groups showed statistically significant decreased levels of TG (P<0.01) (Fig. 4D).
Real-time reverse transcription-polymerase chain reaction showed elevated mRNA expressions of TNF-α, IL-6, and IL-1β in the Western diet group. However, the mRNA expression levels of these inflammatory genes were decreased in the L. acidophilus, L. fermentum, L. paracasei, and L. plantarum groups. The results were not statistically significant, and only a decreasing trend was observed (Fig. 4E).
In the phylum level, the composition of Proteobacteria, Verrucomiccrobia, Actinobacteria, Bacteroidetes, Firmicutes, Deferribacteres, and etc. was different for each group (normal control [3%, 1%, 1%, 52%, 40%, 0%, 0%], Western diet [10%, 6%, 3%, 1%, 71%, 5%, 0%], and L. paracasei + Western diet [3%, 46%, 0%, 3%, 45%, 0%, 1%]) (Fig. 5A). Bacteroidetes and Firmicutes are some of the major phyla of the domain bacteria that dominate the human gut microbiota. Bacteroidetes were dominant in the normal controls (52.1±11.9%) compared with the Western diet group (1.5±1%), and was increased in the L. paracasei + Western diet group (4±3.9%). Firmicutes was increased in the Western diet group (71.7±12.6%) compared to the normal controls (40.5±8%), and was decreased in the L. paracasei + Western diet group (45.2±11.3%) (Fig. 5B). The taxonomic composition of MTP set of all individuals in each group is shown. Bacteroidetes dominated in the normal control group, while Firmicutes dominated in the Western diet group. Firmicutes decreased and Verrucomicroiba increased when L. paracasei was ingested. (Fig. 5C). We measured the differences between the groups using beta diversity for the relationship between microbiome taxonomic profiling, and each group showed a different location (Fig. 5D).
NAFLD can progress to various liver diseases such as cirrhosis, liver failure, and hepatocellular carcinoma in varying proportion of patients [23]. Due to the gut-liver axis, gut microbiome is an important cornerstone in determining the occurrence and progression of chronic liver disease [24]. Recently, human and animal studies have shown the changes of microbiome in the presence and prognosis of chronic liver disease.
There is evidence that cholesterol accumulation contributes to NAFLD, and that lowering cholesterol can be used therapeutically. Also, studies have shown that cholesterol-lowering diet can improve NASH, and adding cholesterol to one’s diet may cause liver inflammation and inflammation of extrahepatic tissue [25,26]. Some studies have shown the effects of L. acidophilus ATCC 4962 and L. fermentum on cholesterol [27,28]. In our study, improvements in total cholesterol levels were shown in the L. acidophilus, L. fermentum, and L. plantarum groups compared to the Western diet control group. Ingestion of Lactobacillus can be a useful therapeutic option for the treatment of NAFLD with high cholesterol. However, there is still a lack of evidence regarding the mechanisms in lowering cholesterol.
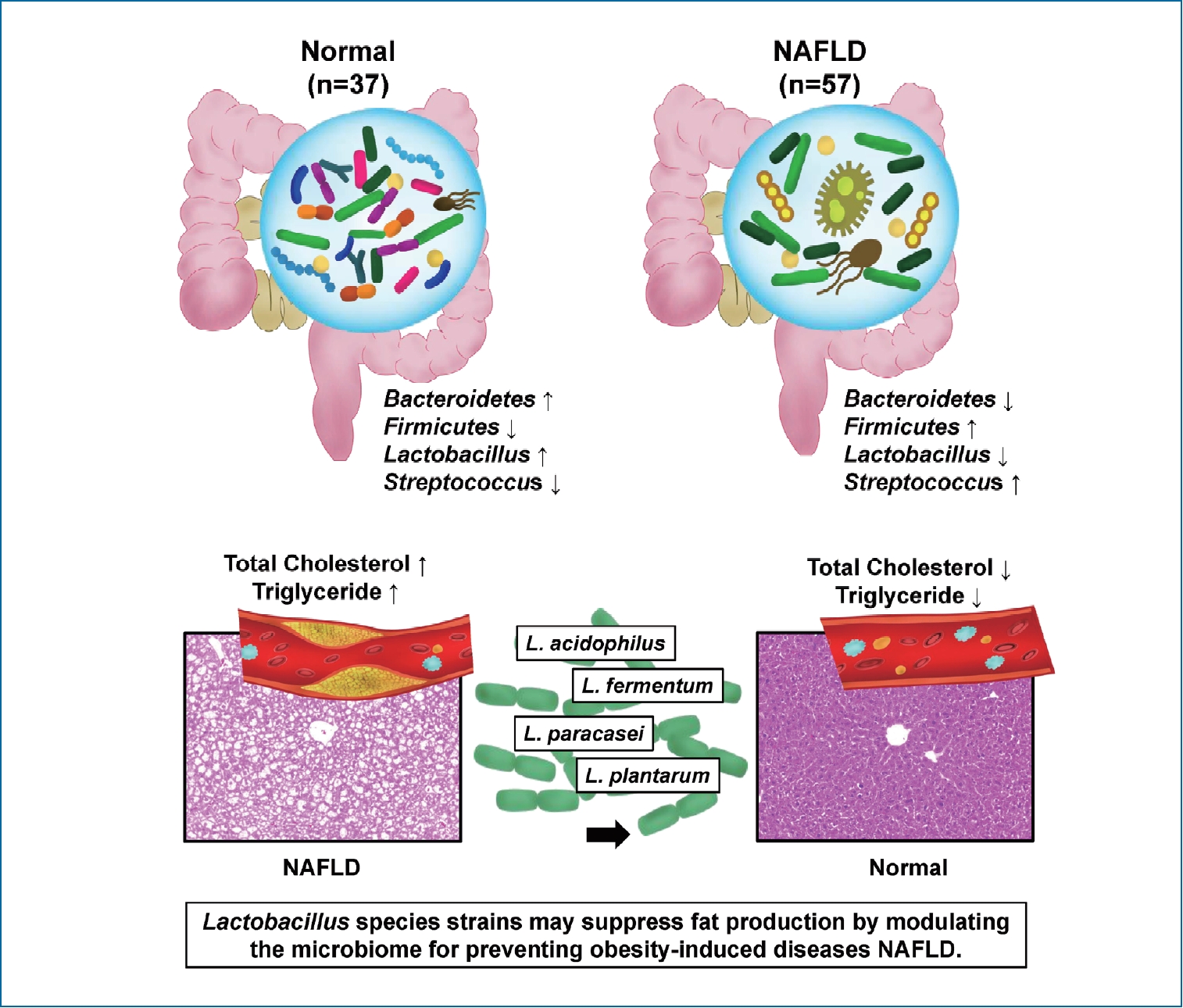
In our study, there was a significant difference in the proportion of phylum level in stool microbes between normal and NAFLD patients. Many studies showed that various factors, including age, genetic, diet, antibiotics, and host immune system can modify the gut microbiome from the normal state [29]. Each person has different composition of gut microbes, which has been used as a health indicator in various situations. Many existing studies have already identified the differences in gut microbiota between healthy people and patients, which are targeted as a treatment goal. According to the results of our clinical analysis, NAFLD patients had a lower rate of Bacteroidetes and a higher rate of Firmicutes compared to the normal control group. As a result, F/B ratio was increased in the NAFLD group [30].
Recent studies have shown the role of gut microbiome in the prevention and treatment of liver disease [31]. The evidence of beneficial role of probiotics has been suggested in the necrotizing enterocolitis, acute infectious diarrhea, acute respiratory infections, antibiotics-related diarrhea, and disorders in infants [32]. A previous study demonstrated that the dysfunction of intestinal barrier and an increase in bacterial translocation to the liver through the gutliver axis play an important role in chronic liver disease [33]. In our results, L. acidophilus, L. plantarum, and L. paracasei supplementation reduced the steatosis grade of liver. Therefore, probiotics might improve the liver histology by modulating dysbiosis and, consequently, attenuate the progression of NAFLD.
Currently, some microbiota can be used as a functional raw material for probiotics, and a total of 21 probiotic strains have been recognized as probiotics by South Korea’s Ministry of Food and Drug Safety, including Lactobacillus, Bifidobacterium, Lactococcus, Enterococcus, and Streptococcus. Among these strains, L. acidophilus, L. fermentum, L. paracasei, and L. plantarum were selected as a treatment option in this study. The effect of probiotics on liver disease has been experimented by several studies [16,34].
In previous studies, L. acidophilus NS1 significantly reduced obesity and liver lipid accumulation with concomitant improvement in insulin sensitivity in subjects consuming a high-fat diet [35]. In the high fat-diet model, L. acidophilus ATCC 43121 intake affected the total cholesterol metabolism and increased the excretion of sterols in feces [36]. Previous studies have indicated that L. acidophilus is a beneficial probiotic for lowering cholesterol and improving liver lipid metabolism [37]. Our animal study has also shown reduced fat accumulation in the liver and notably lowered total cholesterol and TG levels by ingestion of L. acidophilus. Overall, probiotics have an effective role in reducing liver steatosis in NAFLD.
L. fermentum MJM60397 strain has already been proposed as a potential probiotic that increases the excretion of bile acids from feces and lowers serum cholesterol levels [38]. Oral administration of L. fermentum LA12 improves liver function and hepatic steatosis, and also helps restore epithelial barrier function, reducing endotoxin leakage from the blood [39]. In our study, ingestion of L. fermentum was not statistically meaningless in terms of improved steatosis, but total cholesterol and TG levels were significantly lowered. Therefore, there is a discrepancy in the effect of L. fermentum, and further studies on the mechanism of L. fermentum on NAFLD are required.
L. paracasei F19 was shown to control triglyceride deposition into adipocytes [40]. B. longum BORI and L. paracsei CH88 mixture suppressed weight gain and lipid deposition in the liver. The adipocyte was significantly decreased from the mixture group fed with high fat [41]. As a result of our animal experiments, L. paracasei lowered the TG level in liver tissue, and confirmed that it improves steatosis in the liver tissue.
In a previous study, patients with hypercholesterolemia who consumed L. plantarum for 12 weeks significantly reduced their cholesterol levels by 13.6%. L. plantarum has a better effect on patients with high cholesterol levels in proportion to their cardiovascular risk [42]. In the NAFLD model, L. plantarum NCU116 intake for 5 weeks was found to reduce the level of fat accumulation in the liver [43]. In our study, L. plantarum also lowered the total cholesterol levels and improved steatosis levels in the liver.
Previous studies have shown that the Western diet contributes to changes in the composition of gut microbiome [7]. This study also confirmed that the Western diet changed the composition of animal gut microbiome by stool analysis. Bacteroidetes are known to be healthy gut microbes [44]. In our animal study, when L. paracasei was administered to NAFLD animal group, the proportion of Bacteroidetes increased compared to the Western diet group. This suggests that the intake of Lactobacillus ameliorate to improve the NAFLD by modulating gut microbiota.
Overall, according to the results of our animal experiment, the strains decreased the body weight along with lower serum total cholesterol, triglyceride, and steatosis levels. Pro-inflammatory cytokines were decreased and improved, but only showed a decreasing trend, probably as a result of not being given the Western diet for a long time. It is considered a one-shot state in which only fat is formed among the two-shot theory of fatty liver progression. Cytokines are known to play an important role in coordinating the production of many other mediators associated with chronic liver disease and affecting all liver cell types [45]. Further research is needed to determine whether to prolong the intake period of the Western diet and to strengthen the concentration of strains.
The purpose of this study was to analyze the intestinal bacteria of normal and NAFLD subjects based on the fecal results of clinical patients, as well as to confirm the difference and determine how the strains showing the difference affect the fatty liver. In conclusion, this study suggests that gut-microbiota-liver interaction plays an important role in NAFLD. It also showed that probiotics reduced liver steatosis and cholesterol in NAFLD.
Many studies are being conducted to identify the role of probiotics in various diseases. However, there is insufficient research on how probiotics affect diseases. Therefore, an analysis of animal experiments based on the human gut microbiome is needed to elucidate the underlying mechanisms. Microbiome identifies the effects of metabolic disorders in the gut microbiome on insulin resistance [46]. Our study was limited in that we did not consider this point; therefore, future studies will need to select bacteria that show therapeutic efficacy in diseases and perform evaluation of insulin resistance. Nevertheless, our results showed that L. acidophilus, L. fermentum, and L. plantarum led to improvement in cholesterol. Also, L. acidophilus, L. paracasei, and L. plantarum were able to confirm efficacy in steatosis grade.
The Bacteroides species account for a much larger proportion of the composition in mice, and the gut microbiome population varies between humans and mice [47]. Moreover, differences, such as the propensity for rodents to eat feces, can have a significant impact on studies involving gut microbiome [48]. Although these limitations exist in this study, it has demonstrated a trend that when the disease worsens, the representative beneficial bacteria decrease and harmful bacteria increase. Therefore, it must be complemented, in terms of humanized animal research, for rapid progress of clinical application. Further studies are needed to analyze microbial communities in animal models using humanized or sterile mice. Our study suggests modulating the gut microbiome to affect the liver disease so that selective bacteria can be used for treatment, and we believe this can be used in a customized medical service.
ACKNOWLEDGMENTS
This research was supported by Hallym University Research Fund, Korea National Research Foundation (NRF2018M3A9F3020956 and NRF-2018M3A9F3020942), Basic Science Research Program (2020R1A6A1A03043026) through the NRF funded by the Ministry of Education, and Hallym University Research Fund 2018 (HURF-2018-67). The experiment was conducted with support from CKD bio, and Chunlab helped with the analysis.
FOOTNOTES
Authors’ contribution
NYL and KTS designed the study; NYL, SHH, BYK, BKK and KTS performed the experiments and collected data; KTS selected the patients, provided the samples; NYL and KTS analysis data and drafting of the manuscript; All other authors assisted the experiments; NYL, MJS and KTS reviewed and refined the manuscript.
Figure 1.
Patient enrollment diagram. Ninety-four patients participated in this experiment. According to their conditions, patients were divided into either normal control or NAFLD with elevated liver enzyme groups. NAFLD, nonalcoholic fatty liver disease; AST, aspartate aminotransferase; ALT, alanine aminotransferase; BMI, body mass index.
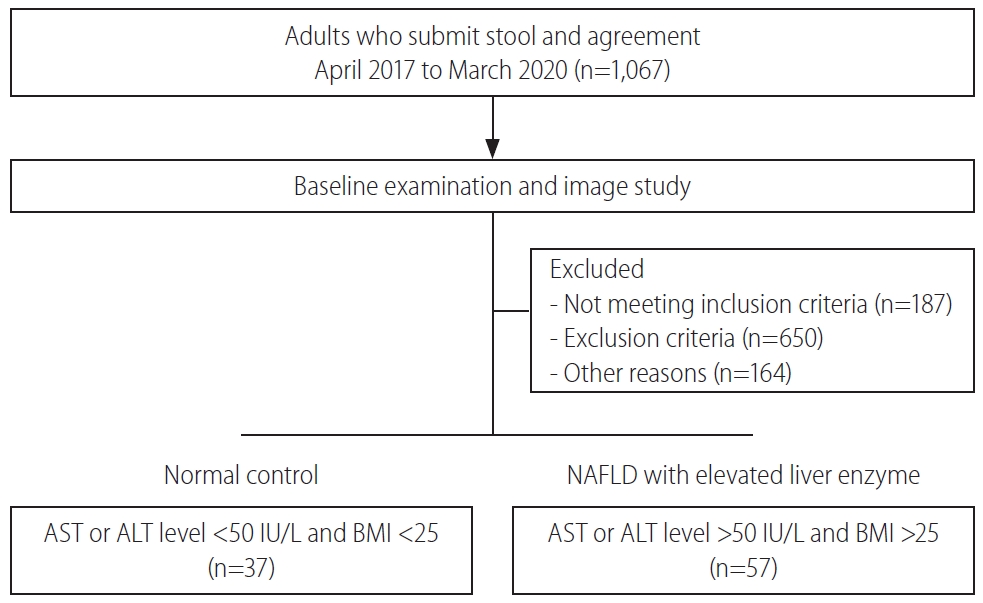
Figure 2.
Human gut microbiome was analyzed for 16s rRNA. (A) Phylum composition of normal controls and NAFLD patients with elevated liver enzymes. (B) The relative abundance of Bacteroidetes and Firmicutes. F/B ratio. (C) Composition of Lactobacillales family in normal controls and NAFLD patients. (D) Composition of Lactobacillus genus in normal controls and NAFLD patients. (E) Lactobacillus species composition. NAFLD, nonalcoholic fatty liver disease; ETC, et cetera; F/B, Firmicutes/Bacteroidetes; NS, not significant. *P<0.05. †P<0.01.
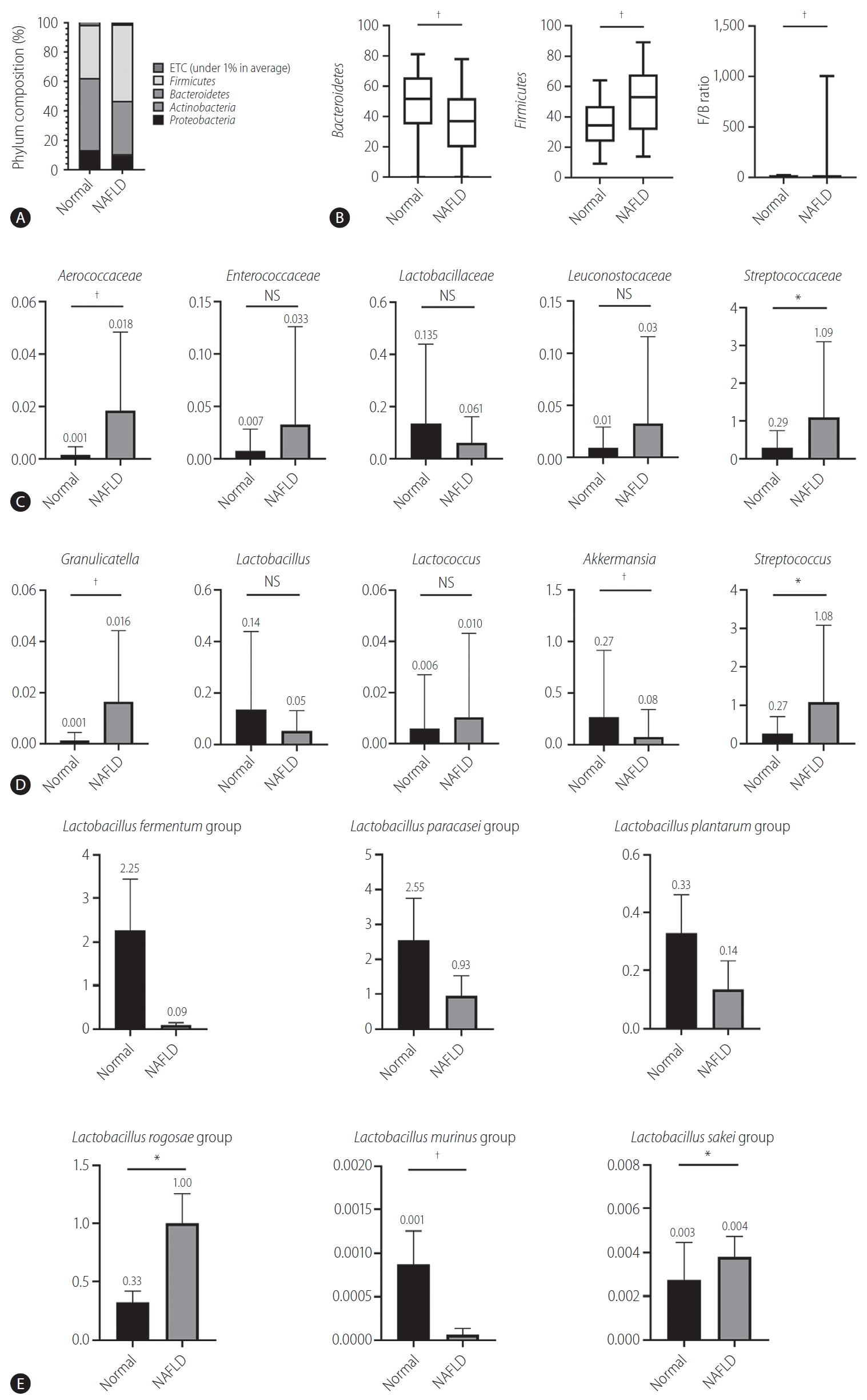
Figure 3.
Result of NAFLD model induced by Western diet. (A) Flow chart of the animal experiment. (B) Gross specimen of mice liver. (C) Effect of strains on body weight gain. (D) Body weight, Liver weight, and L/B ratio in each group. L/B, liver/body; NAFLD, nonalcoholic fatty liver disease. *P<0.05. †P<0.01.
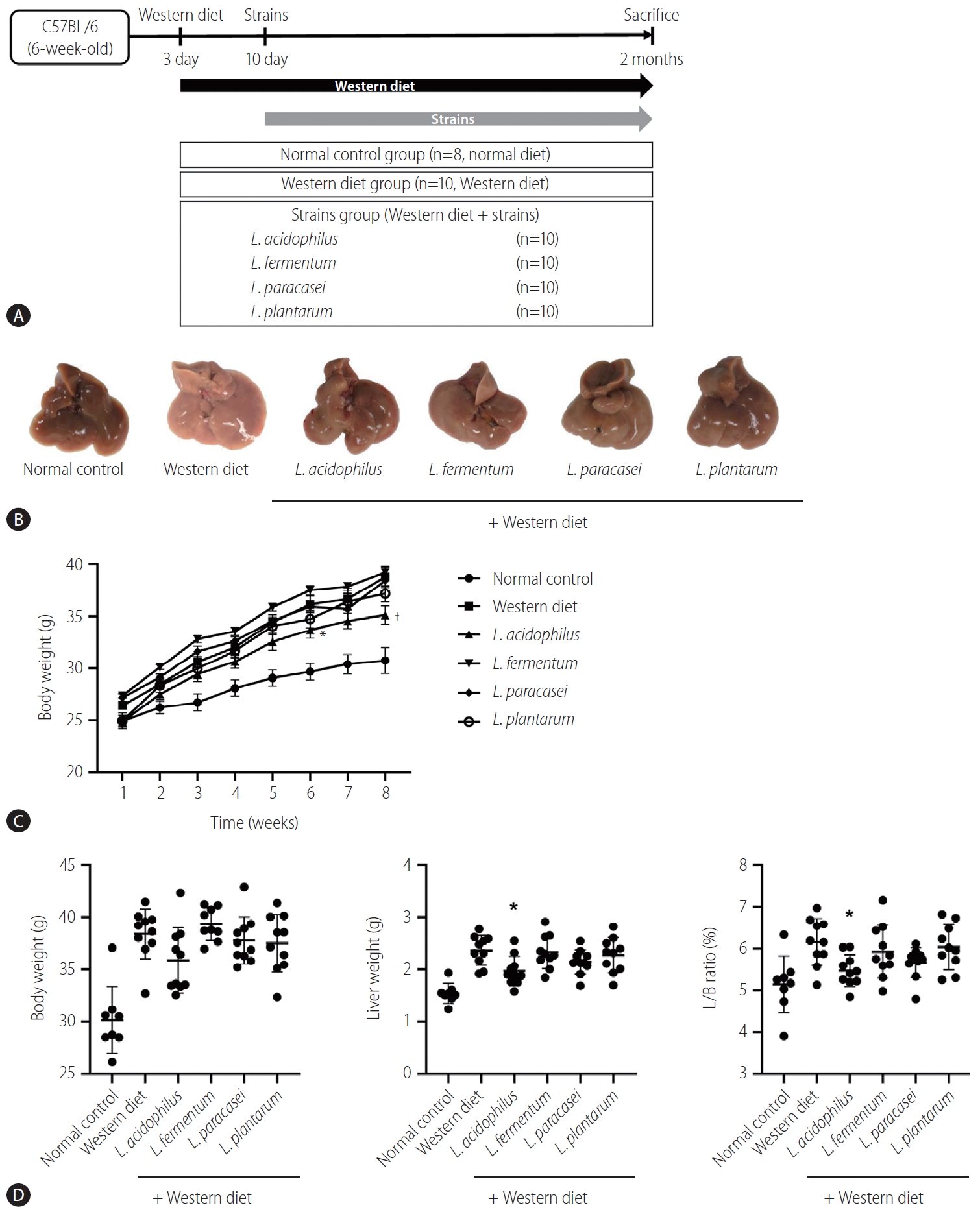
Figure 4.
Effects of strains on serum cholesterol level and steatosis of the liver. (A) Effect of strains on liver function test by serum biochemistry analysis. (B) Hematoxylin and eosin staining showed steatosis grade. There were large fat vacuoles displacing the nuclei to the edges of the cells (Western diet). L. acidophilus, L. fermentum, L. paracasei, and L. plantarum groups showed minimal to mild mixed macro/microvesicular steatosis (×200). (C) Steatosis grade in the liver. (D) Triglyceride level in the liver tissue. (E) mRNA expression of the liver (n=6). T-CHOL, total cholesterol; AST, aspartate aminotransferase; ALT, alanine aminotransferase; TNF, tumor necrosis factor; IL, interleukin. *P<0.05. †P<0.01.

Figure 5.
Animal stool analysis. (A) Phylum composition of animal stool in normal control, Western diet, and L. paracasei fed with Western diet groups. (B) Bacteroidetes and Firmicutes ratio in each group. (C) Taxonomic compositions of MTP sets in each group (n=3). (D) Analytics for beta diversity for the relationship between microbiome taxonomic profiling (principal coordinates analysis, generalized UniFrac, species, include unclassified OTUs). ETC, et cetera; F/B, Firmicutes/Bacteroidetes; PC, principal coordinate analysis; MTP, multi tough point; OTUs, operational taxonominc unit. *P<0.05.
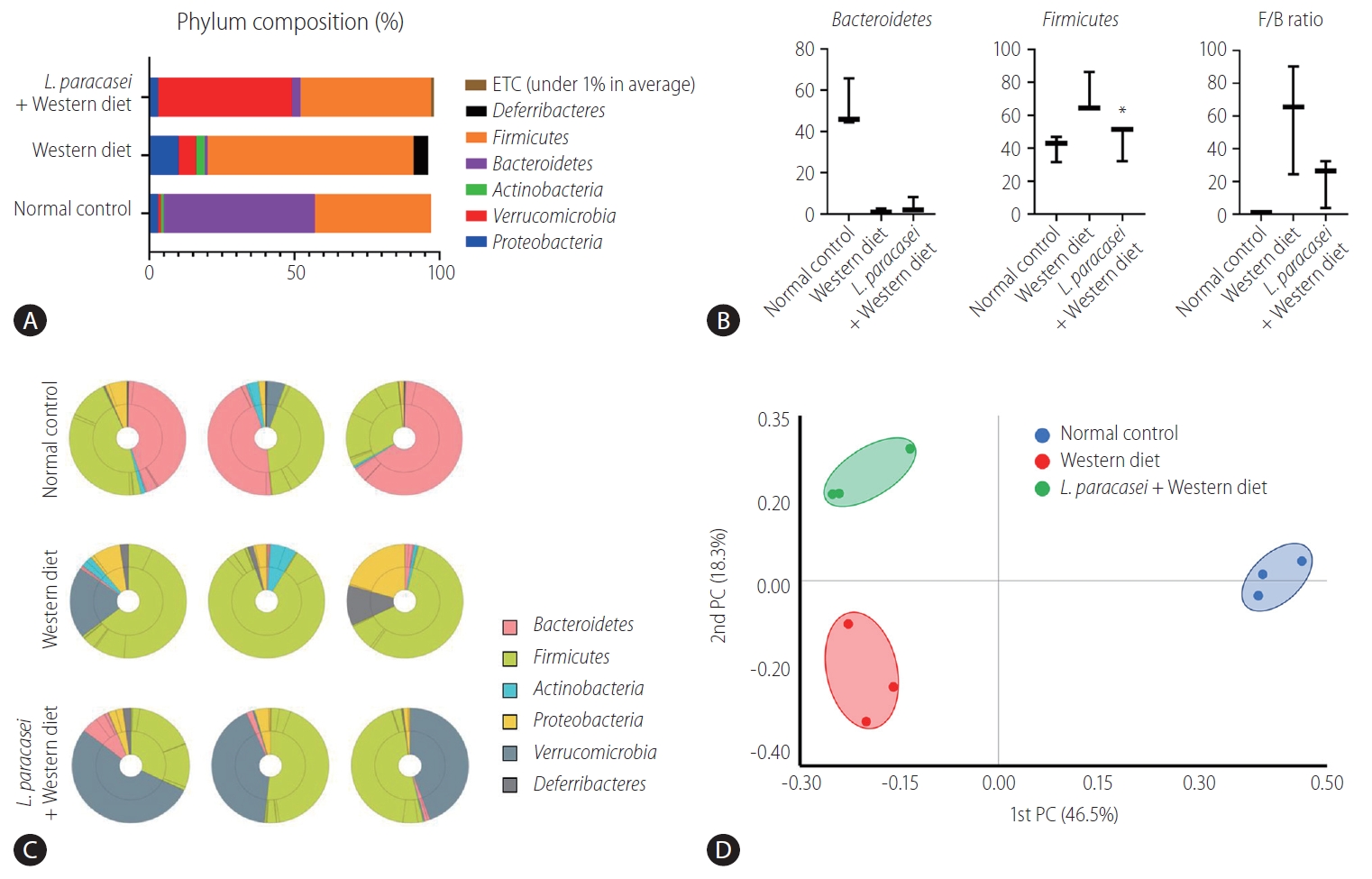
Table 1.
Baseline characteristics of patients
Table 2.
Information of used probiotics strains
| Strain | Number of bacteria (CFU/g) | Characteristics | Known roles in disease |
|---|---|---|---|
| L. acidophilus | 2.80E+11 | Gram-positive | Prevent and reduce diarrhea [49] |
| Reduce total cholesterol [35] | |||
| L. fermentum | 1.50E+11 | Gram-positive | Ameliorate colitis [43] |
| Increase bile acid excretion [37] | |||
| L. paracasei | 4.80E+11 | Gram-positive | Decrease body fat [41] |
| Inhibit lipid deposition [41] | |||
| L. plantarum | 1.00E+12 | Gram-positive | Fortifies the intestinal barrier [50] |
| Decrease cholesterol levels [42] |
Abbreviations
ALT
alanine aminotransferase
AST
aspartate aminotransferase
BMI
body mass index
CFU
colony-forming units
F/B
Firmicutes/Bacteroidetes
FBS
fasting blood sugar
IL
interleukin
MTP
multi touch point
MRS
de Man, Rogosa and Sharpe
NAFLD
nonalcoholic fatty liver disease
NCBI
National Center for Biotechnology Information
PCR
polymerase chain reaction
SRA
Sequence Read Archive
TG
triglyceride
TNF
tumor necrosis factor
γGT
gamma glutamyl transferase
REFERENCES
1. Ascha MS, Hanouneh IA, Lopez R, Tamimi TA, Feldstein AF, Zein NN. The incidence and risk factors of hepatocellular carcinoma in patients with nonalcoholic steatohepatitis. Hepatology 2010;51:1972-1978.


2. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59-65.



3. Cavallari JF, Schertzer JD. Intestinal microbiota contributes to energy balance, metabolic inflammation, and insulin resistance in obesity. J Obes Metab Syndr 2017;26:161-171.



4. Aragonès G, González-García S, Aguilar C, Richart C, Auguet T. Gut microbiota-derived mediators as potential markers in nonalcoholic fatty liver disease. Biomed Res Int 2019;2019:8507583.


5. Takiishi T, Fenero CIM, Câmara NOS. Intestinal barrier and gut microbiota: shaping our immune responses throughout life. Tissue Barriers 2017;5:e1373208.



7. Lee NY, Yoon SJ, Han DH, Gupta H, Youn GS, Shin MJ, et al. Lactobacillus and Pediococcus ameliorate progression of non-alcoholic fatty liver disease through modulation of the gut microbiome. Gut Microbes 2020;11:882-899.



8. Ohtani N, Kawada N. Role of the gut-liver axis in liver inflammation, fibrosis, and cancer: a special focus on the gut microbiota relationship. Hepatol Commun 2019;3:456-470.



9. Saltzman ET, Palacios T, Thomsen M, Vitetta L. Intestinal microbiome shifts, dysbiosis, inflammation, and non-alcoholic fatty liver disease. Front Microbiol 2018;9:61.



10. Walenbergh SM, Shiri-Sverdlov R. Cholesterol is a significant risk factor for non-alcoholic steatohepatitis. Expert Rev Gastroenterol Hepatol 2015;9:1343-1346.


11. Griffin EE, Ullery JC, Cox BE, Jerome WG. Aggregated LDL and lipid dispersions induce lysosomal cholesteryl ester accumulation in macrophage foam cells. J Lipid Res 2005;46:2052-2060.


12. Baratta F, Pastori D, Del Ben M, Polimeni L, Labbadia G, Di Santo S, et al. Reduced lysosomal acid lipase activity in adult patients with non-alcoholic fatty liver disease. EBioMedicine 2015;2:750-754.



13. Hong YF, Kim H, Kim HR, Gim MG, Chung DK. Different immune regulatory potential of Lactobacillus plantarum and Lactobacillus sakei isolated from Kimchi. J Microbiol Biotechnol 2014;24:1629-1635.


14. Uchinaka A, Azuma N, Mizumoto H, Nakano S, Minamiya M, Yoneda M, et al. Anti-inflammatory effects of heat-killed Lactobacillus plantarum L-137 on cardiac and adipose tissue in rats with metabolic syndrome. Sci Rep 2018;8:8156.




16. Wong VW, Won GL, Chim AM, Chu WC, Yeung DK, Li KC, et al. Treatment of nonalcoholic steatohepatitis with probiotics. A proof-of-concept study. Ann Hepatol 2013;12:256-262.


17. Paek JK, Lee SY. Can gut microbiota modulation be used as a practical treatment for obesity? J Obes Metab Syndr 2018;27:75-77.



18. Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 2017;67:1613-1617.



19. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460-2461.


20. Xie JT, Shao ZH, Vanden Hoek TL, Chang WT, Li J, Mehendale S, et al. Antioxidant effects of ginsenoside Re in cardiomyocytes. Eur J Pharmacol 2006;532:201-207.


21. Nyblom H, Björnsson E, Simrén M, Aldenborg F, Almer S, Olsson R. The AST/ALT ratio as an indicator of cirrhosis in patients with PBC. Liver Int 2006;26:840-845.


22. Bahl MI, Bergström A, Licht TR. Freezing fecal samples prior to DNA extraction affects the Firmicutes to Bacteroidetes ratio determined by downstream quantitative PCR analysis. FEMS Microbiol Lett 2012;329:193-197.


23. Bugianesi E, Leone N, Vanni E, Marchesini G, Brunello F, Carucci P, et al. Expanding the natural history of nonalcoholic steatohepatitis: from cryptogenic cirrhosis to hepatocellular carcinoma. Gastroenterology 2002;123:134-140.


24. Betrapally NS, Gillevet PM, Bajaj JS. Gut microbiome and liver disease. Transl Res 2017;179:49-59.



25. Arguello G, Balboa E, Arrese M, Zanlungo S. Recent insights on the role of cholesterol in non-alcoholic fatty liver disease. Biochim Biophys Acta 2015;1852:1765-1778.


26. Tomita K, Teratani T, Suzuki T, Shimizu M, Sato H, Narimatsu K, et al. Free cholesterol accumulation in hepatic stellate cells: mechanism of liver fibrosis aggravation in nonalcoholic steatohepatitis in mice. Hepatology 2014;59:154-169.


27. Liong MT, Dunshea FR, Shah NP. Effects of a synbiotic containing Lactobacillus acidophilus ATCC 4962 on plasma lipid profiles and morphology of erythrocytes in hypercholesterolaemic pigs on high-and low-fat diets. Br J Nutr 2007;98:736-744.


28. Simons LA, Amansec SG, Conway P. Effect of Lactobacillus fermentum on serum lipids in subjects with elevated serum cholesterol. Nutr Metab Cardiovasc Dis 2006;16:531-535.


29. Tuddenham S, Sears CL. The intestinal microbiome and health. Curr Opin Infect Dis 2015;28:464-470.



30. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 2006;444:1022-1023.



31. Schwenger KJ, Clermont-Dejean N, Allard JP. The role of the gut microbiome in chronic liver disease: the clinical evidence revised. JHEP Rep 2019;1:214-226.



32. Liu C, Gong Q, Chen T, Lv J, Feng Z, Liu P, et al. Treatment with 20(S)-ginsenoside Rg3 reverses multidrug resistance in A549/DDP xenograft tumors. Oncol Lett 2018;15:4376-4382.



33. Konturek PC, Harsch IA, Konturek K, Schink M, Konturek T, Neurath MF, et al. Gut–liver axis: how do gut bacteria influence the liver? Med Sci (Basel) 2018;6:79.



34. Ma X, Hua J, Li Z. Probiotics improve high fat diet-induced hepatic steatosis and insulin resistance by increasing hepatic NKT cells. J Hepatol 2008;49:821-830.



35. Park SS, Lee YJ, Song S, Kim B, Kang H, Oh S, et al. Lactobacillus acidophilus NS1 attenuates diet-induced obesity and fatty liver. J Endocrinol 2018;237:87-100.


36. Park YH, Kim JG, Shin YW, Kim SH, Whang KY. Effect of dietary inclusion of Lactobacillus acidophilus ATCC 43121 on cholesterol metabolism in rats. J Microbiol Biotechnol 2007;17:655-662.

37. Song M, Park S, Lee H, Min B, Jung S, Park S, et al. Effect of Lactobacillus acidophilus NS1 on plasma cholesterol levels in diet-induced obese mice. J Dairy Sci 2015;98:1492-1501.


38. Palaniyandi SA, Damodharan K, Suh JW, Yang SH. Probiotic characterization of cholesterol-lowering Lactobacillus fermentum MJM60397. Probiotics Antimicrob Proteins 2020;12:1161-1172.


39. Kim BK, Lee IO, Tan PL, Eor JY, Hwang JK, Kim SH. Protective effect of Lactobacillus fermentum LA12 in an alcohol-induced rat model of alcoholic steatohepatitis. Korean J Food Sci Anim Resour 2017;37:931-939.


40. Aronsson L, Huang Y, Parini P, Korach-André M, Håkansson J, Gustafsson JÅ, et al. Decreased fat storage by Lactobacillus paracasei is associated with increased levels of angiopoietin-like 4 protein (ANGPTL4). PLoS One 2010;5:e13087.



41. Kang D, Li Z, Ji GE. Anti-obesity effects of a mixture of fermented ginseng, Bifidobacterium longum BORI, and Lactobacillus paracasei CH88 in high-fat diet-fed mice. J Microbiol Biotechnol 2018;28:688-696.


42. Fuentes MC, Lajo T, Carrión JM, Cuñé J. Cholesterol-lowering efficacy of Lactobacillus plantarum CECT 7527, 7528 and 7529 in hypercholesterolaemic adults. Br J Nutr 2013;109:1866-1872.


43. Li C, Nie SP, Zhu KX, Ding Q, Li C, Xiong T, et al. Lactobacillus plantarum NCU116 improves liver function, oxidative stress and lipid metabolism in rats with high fat diet induced non-alcoholic fatty liver disease. Food Funct 2014;5:3216-3223.


44. Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019;7:14.



46. Caricilli AM, Saad MJ. The role of gut microbiota on insulin resistance. Nutrients 2013;5:829-851.



47. You JS, Yong JH, Kim GH, Moon S, Nam KT, Ryu JH, et al. Commensal-derived metabolites govern Vibrio cholerae pathogenesis in host intestine. Microbiome 2019;7:132.




48. Bogatyrev SR, Rolando JC, Ismagilov RF. Self-reinoculation with fecal flora changes microbiota density and composition leading to an altered bile-acid profile in the mouse small intestine. Microbiome 2020;8:19.




- TOOLS
-
METRICS

- ORCID iDs
-
Byung Yong Kim

https://orcid.org/0000-0002-4229-8859Ki Tae Suk

https://orcid.org/0000-0002-9206-9245 - Related articles
-
Letter regarding “Non-alcoholic fatty liver disease: Definition and subtypes”2023 July;29(3)
Lean vs. obese phenotypes of nonalcoholic fatty liver disease: similar or different?2023 April;29(2)



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print