| Clin Mol Hepatol > Volume 28(4); 2022 > Article |
|
ABSTRACT
ACKNOWLEDGMENTS
FOOTNOTES
SUPPLEMENTAL MATERIAL
Supplementary┬ĀFigure┬Ā1.
Supplementary┬ĀFigure┬Ā2.
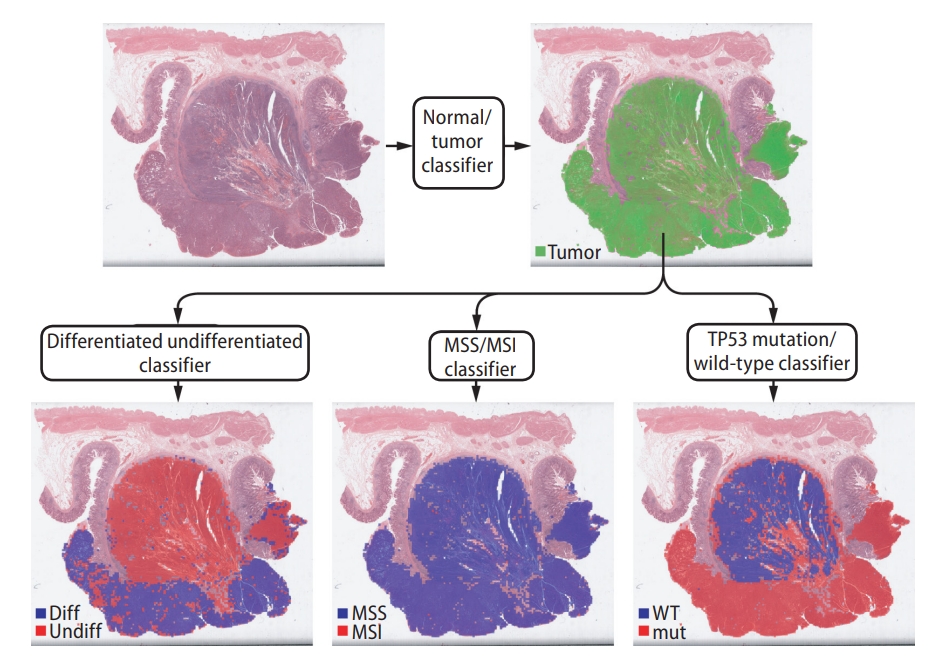
Figure┬Ā1.

Figure┬Ā2.
Table┬Ā1.
| Study | Target (cancer type) | Training cohort | Validation cohort | Neural network | Patch size (magnification) | Performance measure |
|---|---|---|---|---|---|---|
| Schaumberg et al. [8] | SPOP (prostate cancer) | TCGA | MSK-IMPACT | ResNet-50 | 224├Ś224 pixels (N/A) | AUROC: 0.74 |
| Coudray et al. [9] | STK11, KRAS, SETBP1, EGFR, FAT1, and TP53 (lung cancer) | TCGA | NYU Langone Medical Center | Inception v3 | 512├Ś512 pixels (20├Ś) | AUROC: 0.674ŌĆō0.845 |
| Kim et al. [10] | BRAF and NRAS (melanoma) | NYU Langone Health | TCGA | Inception v3 | 229├Ś229 pixels (20├Ś) | AUROC: 0.83 (BRAF) and 0.92 (NRAS) |
| Tsou and Wu [11] | BRAF and RAS (thyroid cancer) | TCGA | None | Inception v3 | 512├Ś512 pixels (5├Ś) | AUROC: 0.951 |
| Chen et al. [12] | CTNNB1, FMN2, TP53, and ZFX4 (liver cancer) | TCGA | SRRSH | Inception v3 | 256├Ś256 pixels (20├Ś) | AUROC: 0.71ŌĆō0.89 |
| Liu et al. [13] | IDH (glioma) | TCGA+YUH | None | ResNet-50 | 256├Ś256 pixels (N/A) | AUROC: 0.927 |
| Fu et al. [14] | 151 gene:cancer pairs (various cancers) | TCGA | METABRIC | Inception v4 | 512├Ś512 pixels (20├Ś) | AUROC: 0.098ŌĆō0.972 |
| Kather et al. [15] | Mutations with a prevalence above 2% (various cancers) | TCGA | None | ShuffleNet | 512├Ś512 pixels (20├Ś) | AUROC: 0.55ŌĆō0.8 |
| Noorbakhsh et al. [17] | TP53 (various cancers) | TCGA | None | Inception v3 | 512├Ś512 pixels (20├Ś) | AUROC: 0.65ŌĆō0.80 |
| Jang et al. [16] | APC, KRAS, PIK3CA, SMAD4, and TP53 (colorectal cancer) | TCGA | SSMH | Inception v3 | 360├Ś360 pixels (20├Ś) | AUROC: 0.645ŌĆō0.809 |
| Yang et al. [19] | DNMT3A, EGFR, PBRM1, STK11, and TP53 (lung cancer) | TCGA | None | ResNet | 512├Ś512 pixels (N/A) | AUROC: 0.71ŌĆō0.87 |
| Loeffler et al. [20] | FGFR3 (bladder cancer) | TCGA | Aachen University | ShuffleNet | 512├Ś512 pixels (20├Ś) | AUROC: 0.701 |
| Jang et al. [21] | CDH1, ERBB2, KRAS, PIK3CA, and TP53 (gastric cancer) | TCGA | SSMH | Inception v3 | 360├Ś360 pixels (20├Ś) | AUROC: 0.661ŌĆō0.862 |
Because some studies resized or cropped the input image patches during preprocessing, the final patch size and magnification are presented in the table. In some cases, the magnification was not clearly specified and thus noted as (N/A). The area under the receiver operating characteristics curves (AUROCs) are for the held-out test sets from the training datasets. If it is not provided, AUROCs for external validation cohort are presented.
TCGA, The Cancer Genome Atlas; MSK-IMPACT, model performed well with the external validation cohort; N/A, not applicable; AUROC, area under the receiver operating characteristics curve; NYU, New York University; SRRSH, Sir Run-Run Shaw Hospital; YUH, Yeditepe University Hospital; METABRIC, Molecular Taxonomy of Breast Cancer International Consortium; SSMH, Seoul St. MaryŌĆÖs Hospital.
Table┬Ā2.
| Study | Target (cancer type) | Training cohort | Validation cohort | Neural network | Patch size(magnification) | Performance measure |
|---|---|---|---|---|---|---|
| Kather et al. [26] | Microsatellite instability (gastrointestinal cancer) | TCGA | DACHS (colorectal), KCCH (gastric) | ResNet-18 | 512├Ś512 pixels (20├Ś) | AUROC: 0.77ŌĆō0.84 |
| Cao et al. [27] | Microsatellite instability (colorectal cancer) | TCGA | Asian CRC cohort | ResNet-18 | 224├Ś224 pixels (20├Ś) | AUROC: 0.8848 |
| Echle et al. [28] | Microsatellite instability (colorectal cancer) | TCGA+DACHS+QUASAR+NLCS | YCR-BCIP | ShuffleNet | 512├Ś512 pixels (20├Ś) | AUROC: 0.92 |
| Wang et al. [29] | Microsatellite instability (endometrial cancer) | TCGA | None | ResNet-18 | 512├Ś512 pixels (20├Ś) | AUROC: 0.73 |
| Krause et al. [30] | Microsatellite instability (colorectal cancer) | TCGA+NLCS | None | ShuffleNet | 512├Ś512 pixels (20├Ś) | AUROC: 0.742ŌĆō0.777 |
| Yamashita et al. [31] | Microsatellite instability (colorectal cancer) | SUMC | TCGA | MobileNet v2 | 224├Ś224 pixels (about 10├Ś) | AUROC: 0.931 |
| Lee et al. [32] | Microsatellite instability (colorectal cancer) | TCGA | SSMH | Inception v3 | 360├Ś360 pixels (20├Ś) | AUROC: 0.861ŌĆō0.942 |
TCGA, The Cancer Genome Atlas; DACHS, Darmkrebs Chancen der Verh├╝tung durch Screening; KCCH, Kanagawa Cancer Center Hospital; AUROC, area under the receiver operating characteristics curve; CRC, colorectal cancer; QUASAR, Quick and Simple and Reliable trial; NLCS, Netherlands Cohort Study; YCR-BCIP, Yorkshire Cancer Research Bowel Cancer Improvement Programme; SUMC, Stanford University Medical Center; SSMH, Seoul St. MaryŌĆÖs Hospital.
Table┬Ā3.
| Study | Target (cancer type) | Training cohort | Validation cohort | Neural network | Patch size(magnification) | Performance measure |
|---|---|---|---|---|---|---|
| Xu et al. [35] | Tumor mutational burden (bladder cancer) | TCGA | None | Xception | 1,024├Ś1,024 pixels (20├Ś) | AUROC: 0.75 |
| Jain and Massoud [36] | Tumor mutational burden (lung cancer) | TCGA | None | Inception v3 | 512├Ś512 pixels (5├Ś, 10├Ś, 20├Ś) | AUROC: 0.92 |
| Xu et al. [37] | Tumor mutational burden (bladder and lung cancers) | TCGA | None | Xception | 512├Ś512 pixels (20├Ś) | AUROC: 0.742ŌĆō0.752 |
| Shimada et al. [38] | Tumor mutational burden (colorectal cancer) | TCGA+Japanese CRC cohort | None | Inception v3 | 300├Ś300 pixels (N/A) | AUROC: 0.934 |
| Sadhwani et al. [39] | Tumor mutational burden (lung cancer) | TCGA | None | Inception v3 | 512├Ś512 pixels (10├Ś) | AUROC: 0.71 |
Table┬Ā4.
| Study | Target (cancer type) | Training cohort | Validation cohort | Neural network | Patch size(magnification) | Performance measure |
|---|---|---|---|---|---|---|
| Couture et al. [44] | Molecular subtypes (breast cancer) | CBCS3 | None | VGG16 | 800├Ś800 pixels (20├Ś) | Accuracy: 77% |
| Kather et al. [15] | Molecular subtypes (breast, colorectal, gastric, and lung cancers) | TCGA | None | ShuffleNet | 512├Ś512 pixels (20├Ś) | AUROC: 0.24ŌĆō0.86 |
| Jaber et al. [45] | Molecular subtypes (breast cancer) | TCGA | None | Inception v3 | 400├Ś400 pixels (5├Ś, 10├Ś, 20├Ś) | Accuracy: 67.27% |
| Hong et al. [46] | Molecular subtypes (endometrial cancer) | TCGA+TCIA | None | InceptionResNet | 299├Ś299 pixels (2.5├Ś, 5├Ś, 10├Ś) | AUROC: 0.827ŌĆō0.934 |
| Sirinukunwattana et al. [47] | Molecular subtypes (colorectal cancer) | FOCUS | TCGA+GRAMPIAN | Inception v3 | 299├Ś299 pixels (3├Ś, 12├Ś) | AUROC: 0.86ŌĆō0.92 |
| Yu et al. [48] | Molecular subtypes (lung cancer) | TCGA | ICGC | VGGNet | 224├Ś224 pixels (about 5├Ś) | AUROC: 0.7ŌĆō0.892 |
| Woerl et al. [49] | Molecular subtypes (bladder cancer) | TCGA | CCC-EMN | ResNet-50 | 512├Ś512 pixels (40├Ś) | AUROC: 0.76ŌĆō0.89 |
Table┬Ā5.
| Study | Target (cancer type) | Training cohort | Validation cohort | Neural network | Patch size(magnification) | Performance measure |
|---|---|---|---|---|---|---|
| Couture et al. [44] | Estrogen receptor (breast cancer) | CBCS3 | None | VGG-16 | 800├Ś800 pixels (20├Ś) | Accuracy: 84% |
| Sha et al. [56] | PD-L1 status (lung cancer) | Own cohort (Tempus Labs Chicago) | None | ResNet-18 | 446├Ś446 and 32├Ś32 pixels (10├Ś) | AUROC: 0.80 |
| Rawat et al. [57] | Estrogen receptor, progesterone receptor, HER2 (breast cancer) | TCGA | ABCTB | ResNet-34 | 224├Ś224 pixels (20├Ś) | AUROC: 0.71ŌĆō0.88 |
| Naik et al. [58] | Estrogen receptor, progesterone receptor, HER2 (breast cancer) | TCGA+ABCTB | Multiple centers | ResNet-50 | 256├Ś256 pixels (20├Ś) | AUROC: 0.778ŌĆō0.92 |
| He et al. [59] | Expression of multiple genes (breast cancer) | Own cohort | TCGA | DenseNet-121 | 224├Ś224 pixels (20├Ś) | Correlation coefficient: 0.52 |
| Schmauch et al. [60] | Expression of multiple genes (various cancers) | TCGA | None | ResNet-50 | 224├Ś224 pixels (20├Ś) | Correlation coefficient: 0.47 |
| Levy-Jurgenson et al. [61] | Expression of multiple genes (breast and lung cancer) | TCGA | None | Inception v3 | 512├Ś512 pixels (20├Ś) | AUROC: 0.44ŌĆō0.85 |
Table┬Ā6.
| Study | Target (cancer type) | Training cohort | Validation cohort | Neural network | Patch size (magnification) | Performance measure |
|---|---|---|---|---|---|---|
| Hu et al. [62] | Anti-PD1 response (melanoma and lung cancer) | TCGA | PUCH | Xception | 256├Ś256 pixels (20├Ś) | AUROC: 0.645ŌĆō0.778 |
| Johannet et al. [63] | Immune checkpoint inhibitors response (melanoma) | NYU | Vanderbilt University | Inception v3 | 299├Ś299 pixels (10├Ś or 20├Ś) | AUROC: 0.691ŌĆō0.793 |
| Bychkov et al. [64] | Prognosis (colorectal cancer) | HUCH | None | VGG-16 | 224├Ś224 pixels (40├Ś) | AUROC: 0.69 |
| Mobadersany et al. [65] | Prognosis (glioma) | TCGA | None | VGG-19 | 256├Ś256 pixels (20├Ś) | HarrellŌĆÖs C index: 0.741 |
| Kather et al. [66] | Prognosis (colorectal cancer) | TCGA | DACHS | VGG-19 | 224├Ś224 pixels (20├Ś) | HR: 1.99 |
| Courtiol et al. [67] | Prognosis (mesothelioma) | French MESOBANK | TCGA | ResNet-50 | 224├Ś224 pixels (20├Ś) | C index: 0.643 |
| Skrede et al. [68] | Prognosis (colorectal cancer) | AkUH, AUH, GCCS, VICTOR trial | QUSAR2 trial | MobileNet v2 | 448├Ś448 pixels (10├Ś, 40├Ś) | HR: 3.84 |
| Kulkarni et al. [69] | Recurrence and prognosis (melanoma) | CUIMC, NYUMC, GHS, ISMMS | YSM | Simple 5-layers CNN with RNN | 100├Ś100 pixels (8├Ś) | AUROC: 0.905 |
| HR: 58.7 | ||||||
| Wulczyn et al. [70] | Prognosis (various cancers) | TCGA | None | Similar to MobileNet | 256├Ś256 pixels (N/A) | HR: 1.58 |
| Fu at al. [14] | Prognosis (various cancers) | TCGA | METABRIC | Inception v4 | 512├Ś512 pixels (20├Ś) | C index: 0.53ŌĆō0.67 |
| Saillard et al. [71] | Prognosis (liver cancer) | HMUH | TCGA | ResNet | 224├Ś224 pixels (20├Ś) | C index: 0.78 |
| Wang et al. [72] | Prognosis (stomach cancer) | CHH and JXCH | None | ResNet-50 | 768├Ś768 pixels (20├Ś) | HR: 2.05 |
| Wulczyn et al. [73] | Prognosis (colorectal cancer) | MUG | None | Similar to MobileNet | 256├Ś256 pixels (5├Ś) | AUROC: 0.69ŌĆō0.70 |
| Shim et al. [74] | Recurrence (lung cancer) | 3 hospitals from CUK | 2 hospitals from CUK | ResNet-50 | 224├Ś224 pixels (10├Ś, 40├Ś) | AUROC: 0.76ŌĆō0.77 |
PD1, programmed death 1; TCGA, The Cancer Genome Atlas; PUCH, Peking University Cancer Hospital; AUROC, area under the receiver operating characteristics curve; NYU, New York University; HUCH, Helsinki University Central Hospital; DACHS, Darmkrebs Chancen der Verh├╝tung durch Screening; HR, hazard ratio; MESOBANK, mesothelioma biobank; AkUH, Akershus University Hospital; AUH, Aker University Hospital; GCCS, Gloucester Colorectal Cancer Study; VICTOR, venetoclax with low dose cytarabine for acute myeloid leukaemia; QUSAR2, QUick And Simple And Reliable2; CUIMC, Columbia University Irving Medical Center; NYUMC, New York University Medical Center; GHS, Geisinger Health Systems; ISMMS, Icahn School of Medicine at Mount Sinai; YSM, Yale School of Medicine; CNN, convolutional neural network; RNN, recurrent neural network; N/A, not applicable; METABRIC, Molecular Taxonomy of Breast Cancer International Consortium; HMUH, Henri Mondor University Hospital; CHH, Changhai Hospital; JXCH, Jiangxi Provincial Cancer Hospital; MUG, Medical University of Graz; CUK, Catholic University of Korea.
Abbreviations
REFERENCES
- TOOLS
-
METRICS

- ORCID iDs
-
Hyun-Jong Jang

https://orcid.org/0000-0003-4535-1560 - Related articles



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement1
Supplement1 Print
Print