Non-invasive biomarkers for liver inflammation in non-alcoholic fatty liver disease: present and future
Article information
Abstract
Inflammation is the key driver of liver fibrosis progression in non-alcoholic fatty liver disease (NAFLD). Unfortunately, it is often challenging to assess inflammation in NAFLD due to its dynamic nature and poor correlation with liver biochemical markers. Liver histology keeps its role as the standard tool, yet it is well-known for substantial sampling, intraobserver, and interobserver variability. Serum proinflammatory cytokines and apoptotic markers, namely cytokeratin-18, are well-studied with reasonable accuracy, whereas serum metabolomics and lipidomics have been adopted in some commercially available diagnostic models. Ultrasound and computed tomography imaging techniques are attractive due to their wide availability; yet their accuracies may not be comparable with magnetic resonance imaging-based tools. Machine learning and deep learning models, be they supervised or unsupervised learning, are promising tools to identify various subtypes of NAFLD, including those with dominating liver inflammation, contributing to sustainable care pathways for NAFLD.
INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD) affects over 30% of the general adult population worldwide, and is emerging as an important cause of cirrhosis and hepatocellular carcinoma [1]. Its more active form, non-alcoholic steatohepatitis (NASH), is characterized by the presence of hepatic steatosis, inflammation (both lobular and portal), and hepatocyte ballooning. Assessment of inflammation is important. Although studies have consistently shown that the fibrosis stage [2] has a stronger correlation with adverse liver-related outcomes than features of NASH, inflammation is, after all, the driver of fibrosis progression [3,4]. Moreover, the United States Food and Drug Administration and the European Medicines Agency both accept NASH resolution with no worsening of fibrosis and/or fibrosis improvement with no worsening of NASH as key histological endpoints for conditional approval of new drugs for NASH [5]. Until the regulators accept the use of non-invasive surrogate biomarkers in place of liver biopsy, assessment of inflammation will remain crucial in the drug development process.
With that being said, the assessment of inflammation is difficult. Above all, there is substantial sampling, intraobserver, and interobserver variability in the histological assessment of inflammation and diagnosis of NASH [6]. When paired biopsies are performed to assess the treatment response, errors at each biopsy add up [7]. If the histological reference standard is unreliable, this would underestimate the performance of even an excellent biomarker. Moreover, compared with fibrosis, inflammation changes more rapidly. Therefore, the time interval between liver biopsy and non-invasive test assessment would have a greater impact on the evaluation of inflammation than fibrosis biomarkers. For the same reason, one may expect inflammatory markers to vary over time, and a single-point assessment may not mean much.
In this article, we review blood and imaging biomarkers of inflammation in NAFLD. We also highlight the emerging role of artificial intelligence and machine learning in diagnostics.
LIVER HISTOLOGY
Liver histology remains the standard to assess inflammation and diagnose NASH. Pathologists diagnose NASH based on a global picture that takes into account the degree and pattern of steatosis, inflammation, and hepatocyte ballooning and/or the presence of Mallory-Denk bodies [8]. In 2005, Kleiner and colleagues [9] from the NASH Clinical Research Network proposed the NAFLD activity score, which is the numerical sum of the steatosis grade (0–3), lobular inflammation (0–3), and ballooning (0–2). Later, it was apparent that it is inappropriate to use the score to diagnose NASH, mainly due to the heavy weighting assigned to steatosis [10]. Therefore, a patient can have severe steatosis but mild inflammation, resulting in a high NAFLD activity score but not meeting the pathological diagnosis of NASH. Currently, the NAFLD activity score is mainly used in early-phase clinical trials to evaluate treatment response.
In contrast, Bedossa and colleagues [11] proposed the Steatosis-Activity-Fibrosis score in 2012, thus separating the assessment of steatosis and inflammation. They also developed the Fatty Liver Inhibition of Progression algorithm, which essentially means that one can diagnose NASH when a patient scores 1 or more in steatosis, lobular inflammation, and ballooning [12]. The algorithm has demonstrated a higher degree of interobserver agreement.
One main limitation of the original scores is the relative underweighting of ballooning, which experts agree should be the defining feature of NASH. Besides, complete disappearance of ballooning is uncommon. This explains the very low percentage of patients with NASH resolution in clinical trials, rendering this histological endpoint often useless [13]. Recently, Pai and colleagues [14] proposed to expand the scale of ballooning scoring from 0–2 to 0–4 to increase granularity and reliability of the assessment of NASH.
Other than assessment variability, liver biopsy is also limited by its invasiveness nature, poor patient acceptance, cost, pain, and potential complications [15]. Therefore, it is important to develop non-invasive tests for routine clinical use.
SERUM MARKERS
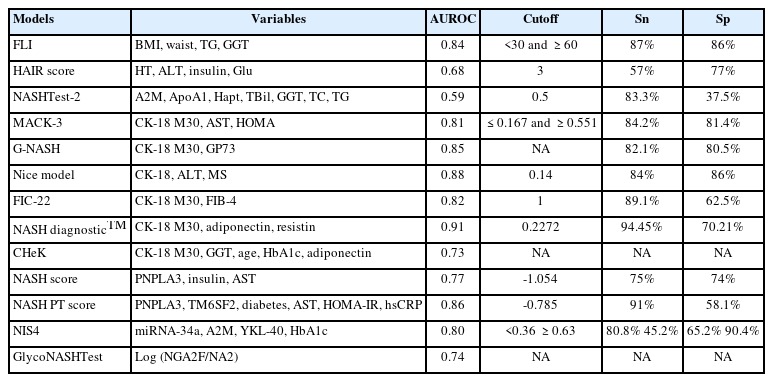
Traditionally, alanine aminotransferase (ALT) and aspartate aminotransferase (AST) have been used in routine clinical practice as biochemical markers of inflammatory damage in hepatocytes, or hepatitis in a simpler term. Unfortunately, a more active form of disease, such as NASH and advanced fibrosis, is often found in NAFLD patients exhibiting normal aminotransferase levels; such levels may even paradoxically decrease in patients with progressive fibrosis [16], suggesting that ALT or AST levels are not reliable in establishing active inflammation in NAFLD. Combining routine clinical parameters is another popular approach; a handful of diagnostic panels were proposed and validated to identify liver inflammation in NASH (Table 1). Most of these models have the benefits of wide availability of parameters included and reasonably good diagnostic accuracy, but specific cut-offs need to be further optimized [17].
Proinflammatory cytokines and apoptotic markers are possible diagnostic biomarkers for patients with NASH. The most evaluated NASH serum biomarker is cytokeratin-18 (CK-18), which is a well-recognized hepatocyte apoptosis product that accounts for about 5% of liver proteins [18]. Two antigens of CK-18, M30 and M65, are of the same protein yet distinctive mechanisms—M30 measures the caspase-cleaved CK-18 revealed during apoptosis, while M65 measures the full-length protein, including both caspase-cleaved and intact CK-18, which is released from cells undergoing necrosis [18]. In general, models with CK-18 perform better than those with solely routine laboratory parameters (Table 1).
Serum metabolomics [19] and lipidomics are also widely studied; pyroglutamic acid, phosphatidylcholine, sphingomyelin, fatty acids, hydroxyeicosatetraenoic acid, glycyrrhetinic acid, taurocholate, and various subtypes of triglycerides levels were incorporated in different models (Table 1) [20]. Some diagnostic models have been commercially available (e.g., by OWL Metabolomics) [21].
While most of the biomarkers and models were derived and validated in a cross-sectional fashion, dedicated studies to evaluate the dynamic change, in particular, the reduction of score after treatment which correlates with inflammation improvement, are much warranted in the era of active development of novel therapeutics for NASH.
ULTRASOUND IMAGING (TABLE 2)
Transabdominal ultrasonography
Conventional B-model ultrasound (US) is the most widely used imaging technique for the non-invasive assessment of NAFLD. Focal steatosis tissue presents brighter than other parenchyma in ultrasound examination because of the increasing attenuation of US waves [22]. US is currently the first-line diagnostic approach for NAFLD suggested by clinical practice guidelines of the European Association for the Study of the Liver due to its low cost, wide availability, and repeatability [23]. In a meta-analysis with 2,815 patients performed on 34 studies, the overall sensitivity of US to detect moderate to severe fatty liver with liver biopsy as a reference standard was 84.8% (95% CI, 79.5–88.9%), specificity was 93.6% (95% CI, 87.2–97.0%) and the AUROC was 0.93 (0.91–0.95) [24]. US has great diagnostic performance for NAFLD.
However, several studies found no correlation between the US characteristics and liver histologic features, including inflammation and ballooning [25,26]. Hamaguchi scoring system was developed based on US findings, including bright liver, and hepatorenal echo contrast (0–3), deep attenuation (0–2), and vessel blurring (0–1). The scoring system further improved the diagnostic performance of NAFLD in obese patients, with an area under the receiver operating characteristic curve (AUROC) of 0.98 [27]. Ultrasonographic fatty liver indicator (US-FLI) is another scoring system ranging from 2–8 based on the intensity of liver or kidney contrast, attenuation of ultrasound beam, vessel blurring, and the visualization of gallbladder wall, diaphragm, and areas of focal sparing. The AUROC of US-FLI for predicting NASH was 0.80 (0.68–0.92), and US-FLI was correlated with lobular inflammation according to Kleiner’s criteria [28]. Hamaguchi score and US-FLI score lack validation in large series of patients, and whether the dynamic change of scores correlates with inflammation progression or improvement needs to be validated in the future.
Vibration-controlled transient elastography
Vibration-controlled transient elastography (VCTE) technique measures the velocity of shear wave through the liver parenchyma, and the velocity is related to the degree of liver tissue stiffness. Controlled attenuation parameter (CAP) captures the attenuation in the amplitude of ultrasound waves to estimate the degree of hepatic steatosis, and it has been available for clinical practice since 2010. Fibroscan 502 Touch was the first VCTE device commercially available with CAP. An examination is considered valid in cases of ≥10 valid liver stiffness measurement (LSM) and CAP, and an interquartile range-to-median ratio of the measurements of ≤0.3 of LSM and CAP [15,29]. According to previous studies, Fibroscan has high accuracy, simplicity, and reproducibility to assess hepatic steatosis and fibrosis [29]. Series of studies have focused on the discriminative ability of CAP and LSM for NASH patients [30,31]. Lee et al. [30] conducted a prospective Korean study based on 183 patients with biopsy-proven NAFLD patients and showed that a cutoff value of 7 kPa for liver stiffness by VCTE can achieve an AUROC of 0.75 (95% confidence interval [CI] 0.68–0.82), a sensitivity of 73.4%, and a specificity of 78.7%. Based on VCTE, they developed a scoring system named “CLA score” using three independent predictors, including CAP value, liver stiffness by VCTE, and ALT level, to identify NASH patients. The CLA score had a significantly higher diagnostic performance than the NAFLD fibrosis score (NFS) (AUROC 0.81 vs. 0.62) [30]. Recently, a randomized phase II drug trial showed that semaglutide in combination with cilofexor groups resulted in the reductions in liver stiffness by VCTE (-2.29 to -3.74 kPa), CAP (-52 to 80 db/m) in 24 weeks, with the improvement in Enhanced Liver Fibrosis score and other liver inflammation biomarkers [32]. The change of liver stiffness over time is also predictors of adverse clinical outcomesTABLE 2[33].
FAST score
FibroScan-AST (FAST) score was a logistic regression-based scoring system for detecting fibrotic NASH, which includes liver stiffness by VCTE, CAP, and AST. The diagnostic performance of FAST score was validated in multiple large global cohorts. AUROCs ranged from 0.74 to 0.95, with sensitivity and specificity up to 1 and 0.86, and NPV ranged from 0.73 to 1. Compared to fibrosis-4 (FIB-4), NFS, and AST to platelet ratio index (APRI), the FAST score had a significantly higher diagnostic performance for fibrotic NASH [34-36]. FAST can be used as a non-invasive tool to screen fibrotic NASH to reduce the number of unnecessary liver biopsies. The relationship between dynamic changes of FAST score and liver inflammation should be explored in the future.
Computed tomography
Computed tomography (CT) uses computer processing of X-ray data of the body to produce images created from the detection of X-rays traversing tissues. Weakening of the X-ray as it passes through the body is a key parameter used to define the brightness of the tissue in the CT image. A healthy liver will appear brighter (i.e., parenchymal hyperdensity) than the spleen in a CT scan. As fat content in the liver increases, its corresponding image will become darker (i.e., parenchymal hypodensity) [37]. CT liver images may be confounded by other factors such as concentration of iron, glycogen, and hematocrit. While CT is widely used to characterize focal liver lesions, in NAFLD patients, CT is more often studied to assess steatosis and fibrosis but not as much for inflammation [38]. Only one retrospective study of 88 NAFLD patients found that non-contrast-enhanced CT texture analysis with a 2-mm filter predicted NASH with accuracy above 90%; yet the accuracy dropped to 60% if a 4-mm filter was used [39]. Other emerging CT techniques, including dual-energy CT, post-processing software, perfusion CT, and photon-counting detector CT, are promising tools that are potentially more accurate to detect inflammation. Currently, CT is not the preferred primary modality to measure liver inflammation given its lack of sensitivity for steatohepatitis and the need for exposure of the subjects to radiation.
MAGNETIC RESONANCE IMAGING (TABLE 3)
LiverMultiScan
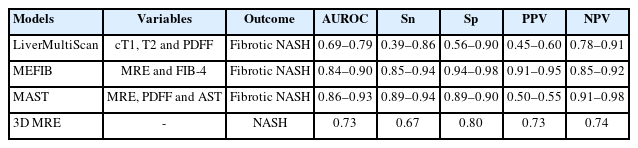
LiverMultiScan (LMS) is an emerging diagnostic tool using multiparametric magnetic resonance imaging (MRI) to quantify liver disease [40]. The technology is comprised of corrected T1 (cT1), T2, and liver fat assessment by advanced MRI. LMS measures the amount of iron in the liver to correct for its effect on T1-cT1, as excess iron in the liver reduces T1 relaxation time and leads to underestimation of liver disease. cT1 correlates with necroinflammation and fibrosis, and may serve as a non-invasive method in NASH. LMS had fewer technical failures, especially compared with ultrasound-based techniques which were less reliable in patients with a higher body mass index. The success rate exceeded 95% in previous clinical studies. One recent pooled study examined the utility of cT1 and proton density fat fraction (PDFF) for identifying NASH and fibrotic NASH [41]. The diagnostic accuracy (AUROC) of cT1 to identify patients with NASH was 0.78 (95% CI, 0.74–0.82), while that for MRI liver fat was 0.78 (95% CI, 0.73–0.82); and when combined cT1 with MRI liver fat, the diagnostic accuracy was 0.82 (95% CI, 0.78–0.85). The diagnostic accuracy of cT1 to identify patients with fibrotic NASH (AUROC [0.78; 95% CI, 0.74–0.82]) was superior to that of MRI liver fat (AUROC [0.69; 95% CI, 0.64–0.74]). There is one ongoing study (NCT03743272) which aims to investigate the repeatability and reproducibility of LMS. Multiparametric MRI has been evaluated to be associated with liver-related clinical outcomes in a cohort of patients with chronic liver disease [42]. Longitudinal change of MRI-PDFF correlated well with the biopsy results, and there was one study evaluated that a 30% relative decline in MRI-PDFF predicted fibrosis regression in NAFLD patients [43,44]TABLE 3.
MEFIB
MEFIB index is a combination of MR elastography and FIB-4 used for the identification of fibrotic NASH [45]. In a validation cohort of the study by Jung et al. [45], the positive predictive value (PPV) exceeded 90% with an AUROC of 0.84 (95% CI, 0.78–0.89). MEFIB was evaluated to have a higher diagnostic accuracy than MAST and FAST score for significant fibrosis as well as fibrotic NASH [46,47]. The MEFIB index had a robust association with liver-related outcome with a hazard ratio of 20.6 (95% CI, 10.4–40.8), and the negative predictive value (NPV) for the outcome reached 99.1% at 5 years [48]. Future studies should explore if the dynamic change of MEFIB index is correlated with liver-related outcomes.
MAST
Given that MRI-PDFF has been shown to be more accurate than VCTE-based CAP in identifying all grades of steatosis in patients with NAFLD, and MR elastography is more accurate than VCTE in detecting liver fibrosis, Noureddin et al. [49] proposed the MAST score based on MRI-PDFF, MR elastography, and AST value. In their validation cohort, the MAST score demonstrated high performance and discrimination (AUROC 0.93, 95% CI 0.88–0.97), which was significantly better compared to the NAFLD fibrosis score, FIB-4 index, and FAST score. However, the MEFIB index showed a higher AUROC, and the PPV and NPV reached 95.3% and 90.1%, respectively, for ruling in and ruling out fibrotic NASH compared with MAST in a head-to-head comparison study [47]. There is still a lack of published studies on the prognostication as well as the dynamic change in fibrosis progression or regression by MAST score.
3D MR elastography
Recently, several studies by Allen et al. [50] from Mayo Clinic evaluated the role of three-dimensional (3D) MR elastography in identifying NASH in patients undergoing bariatric surgery. By combing the 3D MR elastography with MRI-PDFF, the AUROC was 0.73 for the diagnosis of NASH. Additionally, they demonstrated that the 3D MR elastography and MRI-PDFF could detect histologic changes in NASH resolution after bariatric surgery [51]. There are limited studies on the association between 3D MR elastography and liver-related outcomes.
MACHINE LEARNING MODELS
Over the past decade, the advancement of artificial intelligence has led to its numerous applications in hepatology. Artificial intelligence, machine learning, and deep learning can be considered three overlapping domains that use computer programs to mimic functions of human intelligence, including learning, problem solving, classification, and decision making [52]. Particularly, machine learning methods are usually applied for developing diagnostic or predictive models. Machine learning and deep learning algorithms can be supervised or unsupervised. Supervised learning methods occur when a label for the outcome is given in the training data. For example, if we aim to predict the presence of NASH among patients with biopsy-proven NAFLD, the information of whether the patients had NASH needs to be provided to the learning algorithms during training so that the model can distinguish patients with and without NASH based on that. As a result, the learning algorithm can identify combinations and interactions of factors that best separate the two groups of patients and yield an accurate prediction. In contrast, information on the presence and absence of NASH is not provided in unsupervised learning. The purpose of unsupervised learning is to identify several clusters of patients who are similar in terms of data distribution. In other words, patients within the same cluster have similar clinical characteristics, which may represent a certain disease phenotype or subtype.
Common supervised machine learning algorithms examined in identifying inflammation in NAFLD patients, including logistic regression with penalization, decision tree, random forest, support vector machine, and different boosting methods. Regarding the use of covariates, existing literature usually includes laboratory parameters or histological features from liver biopsy for the prediction. Fialoke and colleagues [53] utilized electronic health records from the Optum administrative claim dataset to develop machine learning models for identifying NASH patients from NAFLD patients or healthy patients without NAFLD. In this study, NAFLD and NASH were identified based on diagnosis codes. Supervised machine learning algorithms, including logistic regression, decision tree, random forest, and eXtreme Gradient Boosting (XGBoost), were examined. Temporal mean of laboratory parameters, including ALT, AST, and platelets, together with age, gender, race, and the presence of type 2 diabetes, were included as covariates. The four models yielded satisfactory classification performance with an AUROC of over 0.83 in internal validation (Table 4). This study demonstrated the possibility of using machine learning in identifying NASH in a large group of patients, while the good performance may be due to a more obvious separation between healthy individuals and NASH patients.
The NASHmap is another example of machine learning model for predicting NASH. Docherty and colleagues utilized a biopsy cohort to derive the machine learning models. Similarly, logistic regression, classification and regression trees (a.k.a. decision tree), random forest, and XGBoost were considered. Fourteen clinical and laboratory parameters were included in the models, which yielded AUROCs of around 0.7–0.8. Hemoglobin A1c (HbA1c) was found to be the most predictive covariate, followed by AST and ALT. The models were then externally validated in the Optum dataset and demonstrated comparable AUROC. Slightly reduced performance was observed in reduced models using five parameters, including HbA1c, AST, ALT, total protein, and triglycerides [54]. Moreover, Canbay et al. [55] developed a logistic regression model to distinguish NASH from NAFLD in obese patients, with an AUROC of 0.70 in an independent validation cohort. The logistic model included age, gamma-glutamyl transferase, CK-18 M30, adiponectin, and HbA1c [55]. All of these laboratory-based machine learning models highlighted the importance of HbA1c, AST, and ALT in identifying NASH patients. On the other hand, there is emerging evidence of the difference in the characteristics of lipidomic, glycomic, and hormonal features in patients with NAFLD and NASH due to their strong relationship with metabolic syndrome. Perakakis and colleagues [56] incorporated these omics features into machine learning models including support vector machine, k-nearest neighbor classifier, and random forest. Using 29 features, the machine learning models achieved AUROCs of over 0.95 in selecting patients with NASH from patients with NAFLD or healthy individuals in internal validation (Table 4) [56].
Unsupervised learning can be useful to identify clinically relevant subtypes of NAFLD patients, including those with significant liver inflammation. Using a hierarchical clustering algorithm based on Manhattan distance of similarity, Vandromme and colleagues [57] identified five disease subtypes among NAFLD patients. Some of the subtypes showed evidence of liver inflammation, such as a high proportion of elevated ALT, as well as notable comorbidities, such as diabetes and hypertension.
The presence of lobular inflammation is one of the key histological characteristics of NAFLD activity score (NAS) besides the presence of hepatocyte ballooning and steatosis. Traditional scoring systems, such as the NAFLD activity score, only offer a non-linear and categorical assessment of the disease. Thus, machine learning has a role here to provide quantification of the assessment [58]. Liu and colleagues [58] developed an algorithm to analyze the liver biopsy and quantify different components of the NASH Clinical Research Network (CRN) scoring system. They used special microscopy and image analysis to visualize and quantify inflammation in liver biopsy [58]. The algorithms performed well in a three-center study to predict lobular inflammation and other components of the NASH CRN scoring system (Table 4).
DEEP LEARNING METHODS
Deep learning methods attempt to train deep neural networks for solving complex problems and show more promising prediction results compared to traditional methods based on handcrafted features. Recent deep learning techniques have led to wide applications in healthcare areas [59], and they have been increasingly applied for the prediction and diagnosis of NASH. Popular deep learning approaches include the Convolutional Neural Network (CNN), Graph Neural Network (GNN), and Recurrent Neural Network (RNN). Besides developing sophisticated network architectures to improve prediction accuracy, other important questions in deep learning methods are also explored, such as model interpretability and annotation-efficient learning.
CNN is the most widely used technique of deep learning and has been proved effective in solving many medical problems. CNN achieves better performance when dealing with image-related tasks, such as analyzing CT, MRI, and pathology data. A typical model based on CNN contains a series of layers, including convolution layers, pooling layers, and fully connected layers. In convolution layers, each convolutional neuron only processes data within its receptive field, thus the architecture is ideal for large-scale data such as high-resolution images. NAS is important for diagnosing NASH, and liver biopsy is used for calculating NAS. CNN can be used for quantitative measurement of liver histology and disease monitoring in NASH, and CNN-based methods are proven accurate with strong correlations with expert pathologists and good risk stratification of patients with NASH [60,61]. CT is non-invasive and less expensive compared to liver biopsy, and recent works have proposed to combine the information from CT and pathology data for predicting NAS and fibrosis stage [62]. CNN is first used for feature extraction, and different fusion strategies are proposed to combine these two pieces of information for better prediction performance. Their results showed that combining data from different modalities is beneficial for improving the prediction performance of NAS. To conclude, existing studies have demonstrated that CNNs can automatically learn better features for NASH diagnosis compared to traditional approaches based on manually designed features.
GNN is a rapidly growing field of deep learning that is suitable for processing graph data which contains rich relation information among elements [63]. GNN is able to extract multi-scale localized spatial features by exchanging information between the nodes of graphs, and its key element is pairwise message passing. There is an increasing number of GNN applications, such as electrical health records modeling and synthesizing chemical compounds. GNN is also attracting more attention in pathology data analysis [64], since it learns features that can well-represent the tissue spatial structure. A recent work proposed to study liver biopsy on two histological stains namely Trichrome (TC) and hematoxylin and eosin (H&E) with GNN [65]. The latent embeddings extracted from the graphs were concatenated to predict NAS, and their results showed superiority over competing methods. Graph representation is able to integrate the tissue features from the whole slide image, and deserves further study in the evaluation of tissue biopsies for NASH diagnosis.
RNN can process data with any length, and is a good choice for sequential data processing [66,67]. Electronic health records (EHRs) contain medical time series of laboratory tests, and RNN-based methods can analyze the conditions of patients using these records. Long short-term memory (LSTM) is a representative method of RNN, and its gating mechanism within each LSTM cell is effective to avoid the long-term dependency problem in standard RNNs. Deep learning approaches based on LSTM are utilized to identify patients at risk of developing NASH, and they have shown better performance compared to other competing methods, such as XGBoost [68]. Considering there is a large amount of EHRs available in hospitals, RNN-based methods can work as powerful tools to analyze these existing valuable data for NASH diagnosis.
Even though deep learning methods have achieved great success in solving many medical problems, applying them in clinical practice remains skeptical. However, deep learning methods are often described as “black boxes,” and interpretability is especially important in the medical domain. Some recent works attempted to deal with the interpretability problem of deep learning methods. One promising solution is to incorporate domain knowledge into model design [69]. For example, clinically interpretable features (e.g., nuclei and fat droplets) can be incorporated into NAS prediction. Pathologists normally focus on the nuclei and fat droplet regions for evaluating a liver biopsy image and developing models to mimic the diagnosis process of pathologists is proven effective [70]. Moreover, the success of deep learning models depends on large-scale training data, while collecting such datasets is extremely difficult in the medical domain. Therefore, developing data-efficient deep learning models is important and requires further study for NASH diagnosis; and one possible solution is to fully utilize free-text reports stored in hospital archiving and communication systems [71].
CONCLUSIONS AND PERSPECTIVES
This review summarizes the latest developments in histological and non-invasive assessments of inflammation in NAFLD. In routine clinical practice, non-invasive tests have already largely replaced liver biopsy in the evaluation of patients with NAFLD. However, liver biopsy remains valuable in cases of diagnostic uncertainty, such as uncertain etiology or indeterminate or conflicting non-invasive test results. At present, liver biopsy is still required in late-phase clinical trials for NASH. The limitation of serial liver biopsies to determine NASH resolution has been well-documented. Artificial intelligence-aided assessment of key histological features, including ballooning and fibrosis, has made much progress and should be incorporated into future clinical trials, subject to agreement by the regulators. To the least, artificial intelligence has consistently demonstrated a much higher reproducibility than traditional pathological assessments. Eventually, the aim should be to use non-invasive tests in both clinical trials and routine clinical practice. With a disease that affects over 30% of the population, non-invasive tests are simply the only feasible option if we are to build robust and sustainable clinical care pathways and improve NAFLD management.
Notes
Authors’ contribution
All authors were responsible for the writing plan, content, drafting and critical revision of the manuscript for important intellectual content.
Conflicts of Interest
Terry Yip has served as a speaker and an advisory committee member for Gilead Sciences. Vincent Wong has served as an advisory committee member for AbbVie, Allergan, Echosens, Gilead Sciences, Janssen, Perspectum Diagnostics, Pfizer and Terns, and a speaker for Bristol-Myers Squibb, Echosens, Gilead Sciences and Merck. Grace Wong has served as an advisory committee member for Gilead Sciences and Janssen, as a speaker for Abbott, Abbvie, Bristol-Myers Squibb, Echosens, Furui, Gilead Sciences, Janssen and Roche, and received research grant from Gilead Sciences. The other authors declare that they have no competing interests.
Acknowledgements
This work was supported by the Health and Medical Research Fund (HMRF) of the Food and Health Bureau (Reference no.: 07180216) awarded to Grace Wong.
Abbreviations
ALT
alanine aminotransferase
CI
confidence interval
DM
diabetes mellitus
HCC
hepatocellular carcinoma
NAFLD
non-alcoholic fatty liver disease
CK-18
cytokeratin-18
CT
computed tomography
MRI
magnetic resonance imaging
NASH
non-alcoholic steatohepatitis
SAF
Steatosis-Activity-Fibrosis
FLIP
Fatty Liver Inhibition of Progression
AST
aspartate aminotransferase
HETE
hydroxyeicosatetraenoic acid
US
ultrasound
US-FLI
ultrasonographic fatty liver indicator
VCTE
vibration-controlled transient elastography
CAP
controlled attenuation parameter
LSM
liver stiffness measurement
NFS
NAFLD fibrosis score
FAST
FibroScan-AST
NECT
non-contrast-enhanced CT
DECT
dual-energy CT
pCT
perfusion CT
PCD-CT
photon-counting detector CT
LMS
LiverMultiScan
cT1
corrected T1
PDFF
proton density fat fraction
FIB-4
fibrosis-4
PPV
positive predictive value
NPV
negative predictive value
3D
three-dimensional
CART
classification and regression trees
HbA1c
hemoglobin A1c
NAS
NAFLD activity score
CRN
Clinical Research Network
CNN
Convolutional Neural Network
GNN
Graph Neural Network
RNN
Recurrent Neural Network
EHRs
electronic health records
LSTM
long short-term memory